import pandas as pd
import numpy as np
import seaborn as sns
| Name | Assignment | Score |
|---|---|---|
| Billy | Homework 1 | 94 |
| Sally | Homework 1 | 98 |
| Molly | Homework 1 | 82 |
| Sally | Homework 2 | 47 |
| Student Name | Quarter | Course | Instructor | Recommend? | Expected Grade | Hours Per Week | Comments |
|---|---|---|---|---|---|---|---|
| Billy | SP22 | DSC 80 | Suraj Rampure | No | A- | 14 | I hate this class |
| Billy | SP22 | DSC 40B | Arya Mazumdar | Yes | B+ | 9 | go big O |
| Sally | SP22 | DSC 10 | Janine Tiefenbruck | Yes | A | 11 | babypandas are so cute |
| Molly | SP22 | DSC 80 | Suraj Rampure | Yes | A+ | 2 | I wish there was music in class |
Only instructors can see individual responses. At cape.ucsd.edu, only overall class statistics are visible.
| Quarter | Course | Instructor | Recommend (%) | Expected Grade | Hours Per Week |
|---|---|---|---|---|---|
| SP22 | DSC 80 | Suraj Rampure | 23% | 3.15 (B) | 13.32 |
| SP22 | DSC 40B | Arya Mazumdar | 89% | 3.35 (B+) | 8.54 |
| SP22 | DSC 10 | Janine Tiefenbruck | 94% | 3.45 (B+) | 11.49 |
The university may be interested in looking at CAPEs results by department.
| Quarter | Department | Recommend (%) | Expected Grade | Hours Per Week |
|---|---|---|---|---|
| SP22 | DSC | 91% | 3.01 (B) | 12.29 |
| SP22 | BILD | 85% | 2.78 (C+) | 13.21 |
Prospective students may be interested in comparing course evaluations across different universities.
| University | Recommend (%) | Average GPA | Hours Per Week |
|---|---|---|---|
| UC San Diego | 94% | 3.12 (B) | 42.19 |
| UC Irvine | 89% | 3.15 (B) | 38.44 |
| SDSU | 88% | 2.99 (B-) | 36.89 |
What is the average number of 'Years' for each 'Degree'? Write code that finds the answer as a Series indexed by 'Degree'.
profs = pd.DataFrame(
[['Brad', 'UCB', 8, 'Neuro', 'Orange'],
['Janine', 'UCSD', 7, 'Math', 'Purple'],
['Marina', 'UIC', 6, 'CS', 'Yellow'],
['Justin', 'OSU', 4, 'CS', 'Yellow'],
['Aaron', 'UCB', 4, 'Math', 'Purple'],
['Soohyun', 'UCSD', 1, 'CS', 'Orange'],
['Suraj', 'UCB', 1, 'CS', 'Purple']],
columns=['Name', 'School', 'Years', 'Degree', 'Color']
)
profs
| Name | School | Years | Degree | Color | |
|---|---|---|---|---|---|
| 0 | Brad | UCB | 8 | Neuro | Orange |
| 1 | Janine | UCSD | 7 | Math | Purple |
| 2 | Marina | UIC | 6 | CS | Yellow |
| 3 | Justin | OSU | 4 | CS | Yellow |
| 4 | Aaron | UCB | 4 | Math | Purple |
| 5 | Soohyun | UCSD | 1 | CS | Orange |
| 6 | Suraj | UCB | 1 | CS | Purple |
year_map = {}
for degree in profs['Degree'].unique():
degree_only = profs.loc[profs['Degree'] == degree]
year_map[degree] = degree_only['Years'].mean()
pd.Series(year_map)
Neuro 8.0 Math 5.5 CS 3.0 dtype: float64
For each unique 'Degree', we make a pass through the entire dataset.
Let's try and avoid passing over the dataset repeatedly.
profs
| Name | School | Years | Degree | Color | |
|---|---|---|---|---|---|
| 0 | Brad | UCB | 8 | Neuro | Orange |
| 1 | Janine | UCSD | 7 | Math | Purple |
| 2 | Marina | UIC | 6 | CS | Yellow |
| 3 | Justin | OSU | 4 | CS | Yellow |
| 4 | Aaron | UCB | 4 | Math | Purple |
| 5 | Soohyun | UCSD | 1 | CS | Orange |
| 6 | Suraj | UCB | 1 | CS | Purple |
You can iterate over the rows of a DataFrame using the iterrows method (though you should rarely need to do this):
for idx, row in profs.iterrows():
print(row, '\n')
Name Brad School UCB Years 8 Degree Neuro Color Orange Name: 0, dtype: object Name Janine School UCSD Years 7 Degree Math Color Purple Name: 1, dtype: object Name Marina School UIC Years 6 Degree CS Color Yellow Name: 2, dtype: object Name Justin School OSU Years 4 Degree CS Color Yellow Name: 3, dtype: object Name Aaron School UCB Years 4 Degree Math Color Purple Name: 4, dtype: object Name Soohyun School UCSD Years 1 Degree CS Color Orange Name: 5, dtype: object Name Suraj School UCB Years 1 Degree CS Color Purple Name: 6, dtype: object
year_map = {}
for idx, row in profs.iterrows():
degree = row['Degree']
person_years = row['Years']
if degree in year_map:
year_map[degree] += np.array([1, person_years])
else:
year_map[degree] = np.array([1, person_years])
year_map
{'Neuro': array([1, 8]), 'Math': array([ 2, 11]), 'CS': array([ 4, 12])}
df = pd.DataFrame(year_map, index=['total', 'years'])
df.loc['years'] / df.loc['total']
Neuro 8.0 Math 5.5 CS 3.0 dtype: float64
'Years' for each 'Degree'?profs
| Name | School | Years | Degree | Color | |
|---|---|---|---|---|---|
| 0 | Brad | UCB | 8 | Neuro | Orange |
| 1 | Janine | UCSD | 7 | Math | Purple |
| 2 | Marina | UIC | 6 | CS | Yellow |
| 3 | Justin | OSU | 4 | CS | Yellow |
| 4 | Aaron | UCB | 4 | Math | Purple |
| 5 | Soohyun | UCSD | 1 | CS | Orange |
| 6 | Suraj | UCB | 1 | CS | Purple |
profs.groupby('Degree').mean()
| Years | |
|---|---|
| Degree | |
| CS | 3.0 |
| Math | 5.5 |
| Neuro | 8.0 |
.csv files.groupby method involves three steps: split, apply, and combine.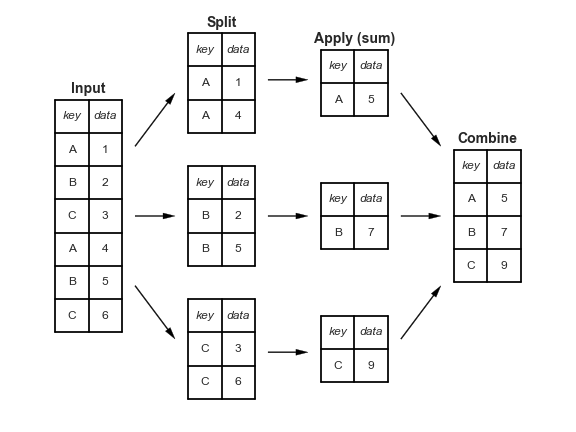
The groupby method can often produce results using just a single pass over the data, updating the sum, mean, count, min, or other aggregate for each group along the way.
groupby is a declarative operation – the user just specifies what computation needs to be done, and pandas figures out how to do it under the hood.
The split-apply-combine pattern can be parallelized to work on multiple computers or threads, by sending computations for each group to different processors.
penguins = sns.load_dataset('penguins').dropna()
penguins.head()
| species | island | bill_length_mm | bill_depth_mm | flipper_length_mm | body_mass_g | sex | |
|---|---|---|---|---|---|---|---|
| 0 | Adelie | Torgersen | 39.1 | 18.7 | 181.0 | 3750.0 | Male |
| 1 | Adelie | Torgersen | 39.5 | 17.4 | 186.0 | 3800.0 | Female |
| 2 | Adelie | Torgersen | 40.3 | 18.0 | 195.0 | 3250.0 | Female |
| 4 | Adelie | Torgersen | 36.7 | 19.3 | 193.0 | 3450.0 | Female |
| 5 | Adelie | Torgersen | 39.3 | 20.6 | 190.0 | 3650.0 | Male |
penguins['species'].value_counts()
Adelie 146 Gentoo 119 Chinstrap 68 Name: species, dtype: int64
penguins['island'].value_counts()
Biscoe 163 Dream 123 Torgersen 47 Name: island, dtype: int64
What is the median bill length?
penguins.groupby('species').median()
| bill_length_mm | bill_depth_mm | flipper_length_mm | body_mass_g | |
|---|---|---|---|---|
| species | ||||
| Adelie | 38.85 | 18.40 | 190.0 | 3700.0 |
| Chinstrap | 49.55 | 18.45 | 196.0 | 3700.0 |
| Gentoo | 47.40 | 15.00 | 216.0 | 5050.0 |
What proportion live on Dream Island?
(
penguins.assign(on_Dream = penguins['island'] == 'Dream')
.groupby('species')
.mean()
)
| bill_length_mm | bill_depth_mm | flipper_length_mm | body_mass_g | on_Dream | |
|---|---|---|---|---|---|
| species | |||||
| Adelie | 38.823973 | 18.347260 | 190.102740 | 3706.164384 | 0.376712 |
| Chinstrap | 48.833824 | 18.420588 | 195.823529 | 3733.088235 | 1.000000 |
| Gentoo | 47.568067 | 14.996639 | 217.235294 | 5092.436975 | 0.000000 |
Now that we understand how to use groupby, let's dive deeper into how it works.
df is a DataFrame, then df.groupby(key) returns a DataFrameGroupBy object.DataFrameGroupBy objects:.groups: a dictionary in which the keys are group names and the values are lists of row labels..get_group(key): a DataFrame with only the values for the given keygroupby works.# Creates one group for each unique value in the species column
penguin_groups = penguins.groupby('species')
penguin_groups
<pandas.core.groupby.generic.DataFrameGroupBy object at 0x7fadf005c190>
penguin_groups.groups
{'Adelie': [0, 1, 2, 4, 5, 6, 7, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35, 36, 37, 38, 39, 40, 41, 42, 43, 44, 45, 46, 48, 49, 50, 51, 52, 53, 54, 55, 56, 57, 58, 59, 60, 61, 62, 63, 64, 65, 66, 67, 68, 69, 70, 71, 72, 73, 74, 75, 76, 77, 78, 79, 80, 81, 82, 83, 84, 85, 86, 87, 88, 89, 90, 91, 92, 93, 94, 95, 96, 97, 98, 99, 100, 101, 102, 103, 104, 105, ...], 'Chinstrap': [152, 153, 154, 155, 156, 157, 158, 159, 160, 161, 162, 163, 164, 165, 166, 167, 168, 169, 170, 171, 172, 173, 174, 175, 176, 177, 178, 179, 180, 181, 182, 183, 184, 185, 186, 187, 188, 189, 190, 191, 192, 193, 194, 195, 196, 197, 198, 199, 200, 201, 202, 203, 204, 205, 206, 207, 208, 209, 210, 211, 212, 213, 214, 215, 216, 217, 218, 219], 'Gentoo': [220, 221, 222, 223, 224, 225, 226, 227, 228, 229, 230, 231, 232, 233, 234, 235, 236, 237, 238, 239, 240, 241, 242, 243, 244, 245, 247, 248, 249, 250, 251, 252, 253, 254, 255, 256, 257, 258, 259, 260, 261, 262, 263, 264, 265, 266, 267, 268, 269, 270, 271, 272, 273, 274, 275, 276, 277, 278, 279, 280, 281, 282, 283, 284, 285, 287, 288, 289, 290, 291, 292, 293, 294, 295, 296, 297, 298, 299, 300, 301, 302, 303, 304, 305, 306, 307, 308, 309, 310, 311, 312, 313, 314, 315, 316, 317, 318, 319, 320, 321, ...]}
penguin_groups.get_group('Chinstrap')
| species | island | bill_length_mm | bill_depth_mm | flipper_length_mm | body_mass_g | sex | |
|---|---|---|---|---|---|---|---|
| 152 | Chinstrap | Dream | 46.5 | 17.9 | 192.0 | 3500.0 | Female |
| 153 | Chinstrap | Dream | 50.0 | 19.5 | 196.0 | 3900.0 | Male |
| 154 | Chinstrap | Dream | 51.3 | 19.2 | 193.0 | 3650.0 | Male |
| 155 | Chinstrap | Dream | 45.4 | 18.7 | 188.0 | 3525.0 | Female |
| 156 | Chinstrap | Dream | 52.7 | 19.8 | 197.0 | 3725.0 | Male |
| ... | ... | ... | ... | ... | ... | ... | ... |
| 215 | Chinstrap | Dream | 55.8 | 19.8 | 207.0 | 4000.0 | Male |
| 216 | Chinstrap | Dream | 43.5 | 18.1 | 202.0 | 3400.0 | Female |
| 217 | Chinstrap | Dream | 49.6 | 18.2 | 193.0 | 3775.0 | Male |
| 218 | Chinstrap | Dream | 50.8 | 19.0 | 210.0 | 4100.0 | Male |
| 219 | Chinstrap | Dream | 50.2 | 18.7 | 198.0 | 3775.0 | Female |
68 rows × 7 columns
# Same as the above
penguins[penguins['species'] == 'Chinstrap']
| species | island | bill_length_mm | bill_depth_mm | flipper_length_mm | body_mass_g | sex | |
|---|---|---|---|---|---|---|---|
| 152 | Chinstrap | Dream | 46.5 | 17.9 | 192.0 | 3500.0 | Female |
| 153 | Chinstrap | Dream | 50.0 | 19.5 | 196.0 | 3900.0 | Male |
| 154 | Chinstrap | Dream | 51.3 | 19.2 | 193.0 | 3650.0 | Male |
| 155 | Chinstrap | Dream | 45.4 | 18.7 | 188.0 | 3525.0 | Female |
| 156 | Chinstrap | Dream | 52.7 | 19.8 | 197.0 | 3725.0 | Male |
| ... | ... | ... | ... | ... | ... | ... | ... |
| 215 | Chinstrap | Dream | 55.8 | 19.8 | 207.0 | 4000.0 | Male |
| 216 | Chinstrap | Dream | 43.5 | 18.1 | 202.0 | 3400.0 | Female |
| 217 | Chinstrap | Dream | 49.6 | 18.2 | 193.0 | 3775.0 | Male |
| 218 | Chinstrap | Dream | 50.8 | 19.0 | 210.0 | 4100.0 | Male |
| 219 | Chinstrap | Dream | 50.2 | 18.7 | 198.0 | 3775.0 | Female |
68 rows × 7 columns
for key, df in penguin_groups:
display(df)
| species | island | bill_length_mm | bill_depth_mm | flipper_length_mm | body_mass_g | sex | |
|---|---|---|---|---|---|---|---|
| 0 | Adelie | Torgersen | 39.1 | 18.7 | 181.0 | 3750.0 | Male |
| 1 | Adelie | Torgersen | 39.5 | 17.4 | 186.0 | 3800.0 | Female |
| 2 | Adelie | Torgersen | 40.3 | 18.0 | 195.0 | 3250.0 | Female |
| 4 | Adelie | Torgersen | 36.7 | 19.3 | 193.0 | 3450.0 | Female |
| 5 | Adelie | Torgersen | 39.3 | 20.6 | 190.0 | 3650.0 | Male |
| ... | ... | ... | ... | ... | ... | ... | ... |
| 147 | Adelie | Dream | 36.6 | 18.4 | 184.0 | 3475.0 | Female |
| 148 | Adelie | Dream | 36.0 | 17.8 | 195.0 | 3450.0 | Female |
| 149 | Adelie | Dream | 37.8 | 18.1 | 193.0 | 3750.0 | Male |
| 150 | Adelie | Dream | 36.0 | 17.1 | 187.0 | 3700.0 | Female |
| 151 | Adelie | Dream | 41.5 | 18.5 | 201.0 | 4000.0 | Male |
146 rows × 7 columns
| species | island | bill_length_mm | bill_depth_mm | flipper_length_mm | body_mass_g | sex | |
|---|---|---|---|---|---|---|---|
| 152 | Chinstrap | Dream | 46.5 | 17.9 | 192.0 | 3500.0 | Female |
| 153 | Chinstrap | Dream | 50.0 | 19.5 | 196.0 | 3900.0 | Male |
| 154 | Chinstrap | Dream | 51.3 | 19.2 | 193.0 | 3650.0 | Male |
| 155 | Chinstrap | Dream | 45.4 | 18.7 | 188.0 | 3525.0 | Female |
| 156 | Chinstrap | Dream | 52.7 | 19.8 | 197.0 | 3725.0 | Male |
| ... | ... | ... | ... | ... | ... | ... | ... |
| 215 | Chinstrap | Dream | 55.8 | 19.8 | 207.0 | 4000.0 | Male |
| 216 | Chinstrap | Dream | 43.5 | 18.1 | 202.0 | 3400.0 | Female |
| 217 | Chinstrap | Dream | 49.6 | 18.2 | 193.0 | 3775.0 | Male |
| 218 | Chinstrap | Dream | 50.8 | 19.0 | 210.0 | 4100.0 | Male |
| 219 | Chinstrap | Dream | 50.2 | 18.7 | 198.0 | 3775.0 | Female |
68 rows × 7 columns
| species | island | bill_length_mm | bill_depth_mm | flipper_length_mm | body_mass_g | sex | |
|---|---|---|---|---|---|---|---|
| 220 | Gentoo | Biscoe | 46.1 | 13.2 | 211.0 | 4500.0 | Female |
| 221 | Gentoo | Biscoe | 50.0 | 16.3 | 230.0 | 5700.0 | Male |
| 222 | Gentoo | Biscoe | 48.7 | 14.1 | 210.0 | 4450.0 | Female |
| 223 | Gentoo | Biscoe | 50.0 | 15.2 | 218.0 | 5700.0 | Male |
| 224 | Gentoo | Biscoe | 47.6 | 14.5 | 215.0 | 5400.0 | Male |
| ... | ... | ... | ... | ... | ... | ... | ... |
| 338 | Gentoo | Biscoe | 47.2 | 13.7 | 214.0 | 4925.0 | Female |
| 340 | Gentoo | Biscoe | 46.8 | 14.3 | 215.0 | 4850.0 | Female |
| 341 | Gentoo | Biscoe | 50.4 | 15.7 | 222.0 | 5750.0 | Male |
| 342 | Gentoo | Biscoe | 45.2 | 14.8 | 212.0 | 5200.0 | Female |
| 343 | Gentoo | Biscoe | 49.9 | 16.1 | 213.0 | 5400.0 | Male |
119 rows × 7 columns
DataFrameGroupBy object, we need to apply some function to each group, and combine the results.DataFrameGroupBy object, e.g. .mean(), .max(), .median(), etc.penguins
| species | island | bill_length_mm | bill_depth_mm | flipper_length_mm | body_mass_g | sex | |
|---|---|---|---|---|---|---|---|
| 0 | Adelie | Torgersen | 39.1 | 18.7 | 181.0 | 3750.0 | Male |
| 1 | Adelie | Torgersen | 39.5 | 17.4 | 186.0 | 3800.0 | Female |
| 2 | Adelie | Torgersen | 40.3 | 18.0 | 195.0 | 3250.0 | Female |
| 4 | Adelie | Torgersen | 36.7 | 19.3 | 193.0 | 3450.0 | Female |
| 5 | Adelie | Torgersen | 39.3 | 20.6 | 190.0 | 3650.0 | Male |
| ... | ... | ... | ... | ... | ... | ... | ... |
| 338 | Gentoo | Biscoe | 47.2 | 13.7 | 214.0 | 4925.0 | Female |
| 340 | Gentoo | Biscoe | 46.8 | 14.3 | 215.0 | 4850.0 | Female |
| 341 | Gentoo | Biscoe | 50.4 | 15.7 | 222.0 | 5750.0 | Male |
| 342 | Gentoo | Biscoe | 45.2 | 14.8 | 212.0 | 5200.0 | Female |
| 343 | Gentoo | Biscoe | 49.9 | 16.1 | 213.0 | 5400.0 | Male |
333 rows × 7 columns
penguin_groups
<pandas.core.groupby.generic.DataFrameGroupBy object at 0x7fadf005c190>
penguin_groups.mean()
| bill_length_mm | bill_depth_mm | flipper_length_mm | body_mass_g | |
|---|---|---|---|---|
| species | ||||
| Adelie | 38.823973 | 18.347260 | 190.102740 | 3706.164384 |
| Chinstrap | 48.833824 | 18.420588 | 195.823529 | 3733.088235 |
| Gentoo | 47.568067 | 14.996639 | 217.235294 | 5092.436975 |
penguin_groups.sum()
| bill_length_mm | bill_depth_mm | flipper_length_mm | body_mass_g | |
|---|---|---|---|---|
| species | ||||
| Adelie | 5668.3 | 2678.7 | 27755.0 | 541100.0 |
| Chinstrap | 3320.7 | 1252.6 | 13316.0 | 253850.0 |
| Gentoo | 5660.6 | 1784.6 | 25851.0 | 606000.0 |
penguin_groups.max()
| island | bill_length_mm | bill_depth_mm | flipper_length_mm | body_mass_g | sex | |
|---|---|---|---|---|---|---|
| species | ||||||
| Adelie | Torgersen | 46.0 | 21.5 | 210.0 | 4775.0 | Male |
| Chinstrap | Dream | 58.0 | 20.8 | 212.0 | 4800.0 | Male |
| Gentoo | Biscoe | 59.6 | 17.3 | 231.0 | 6300.0 | Male |
max and min are defined on strings, while median and mean are not.DataFrameGroupBy objects support [] notation.penguins.groupby('species').median()
| bill_length_mm | bill_depth_mm | flipper_length_mm | body_mass_g | |
|---|---|---|---|---|
| species | ||||
| Adelie | 38.85 | 18.40 | 190.0 | 3700.0 |
| Chinstrap | 49.55 | 18.45 | 196.0 | 3700.0 |
| Gentoo | 47.40 | 15.00 | 216.0 | 5050.0 |
penguins.groupby('species')['bill_length_mm'].median()
species Adelie 38.85 Chinstrap 49.55 Gentoo 47.40 Name: bill_length_mm, dtype: float64
# Gives the same result, but involves wasted effort
# since the other columns had to be aggregated for no reason
penguins.groupby('species').median()['bill_length_mm']
species Adelie 38.85 Chinstrap 49.55 Gentoo 47.40 Name: bill_length_mm, dtype: float64
# Note that this is a SeriesGroupBy object, not a DataFrameGroupBy object!
penguins.groupby('species')['bill_length_mm']
<pandas.core.groupby.generic.SeriesGroupBy object at 0x7fae21144790>
GroupBy methods¶pandas?aggregate method¶DataFrameGroupBy object has a general aggregate method, which aggregates using one or more operations.aggregate; refer to the documentation for a comprehensive list.agg is an alias for aggregate.How many penguins are there of each species, and what is the mean body mass of each species?
penguins.groupby('species')['body_mass_g'].aggregate(['count', 'mean'])
| count | mean | |
|---|---|---|
| species | ||
| Adelie | 146 | 3706.164384 |
| Chinstrap | 68 | 3733.088235 |
| Gentoo | 119 | 5092.436975 |
Note what happens when we don't select a column before aggregating.
# penguins.drop(columns=['island', 'sex']).groupby('species').aggregate(['count', 'mean'])
penguins.groupby('species').aggregate(['count', 'mean'])
/var/folders/pd/w73mdrsj2836_7gp0brr2q7r0000gn/T/ipykernel_19120/3019667236.py:2: FutureWarning: ['island', 'sex'] did not aggregate successfully. If any error is raised this will raise in a future version of pandas. Drop these columns/ops to avoid this warning.
penguins.groupby('species').aggregate(['count', 'mean'])
| bill_length_mm | bill_depth_mm | flipper_length_mm | body_mass_g | |||||
|---|---|---|---|---|---|---|---|---|
| count | mean | count | mean | count | mean | count | mean | |
| species | ||||||||
| Adelie | 146 | 38.823973 | 146 | 18.347260 | 146 | 190.102740 | 146 | 3706.164384 |
| Chinstrap | 68 | 48.833824 | 68 | 18.420588 | 68 | 195.823529 | 68 | 3733.088235 |
| Gentoo | 119 | 47.568067 | 119 | 14.996639 | 119 | 217.235294 | 119 | 5092.436975 |
What is the max bill length of each species, and how many islands is each species found on?
penguins.groupby('species').aggregate({'bill_length_mm': 'max', 'island': 'nunique'})
| bill_length_mm | island | |
|---|---|---|
| species | ||
| Adelie | 46.0 | 3 |
| Chinstrap | 58.0 | 1 |
| Gentoo | 59.6 | 1 |
What is the interquartile range of the body mass of each species?
def IQR(col):
return np.percentile(col, 75) - np.percentile(col, 25)
penguins.groupby('species')['body_mass_g'].aggregate(IQR)
species Adelie 637.5 Chinstrap 462.5 Gentoo 800.0 Name: body_mass_g, dtype: float64
transform method¶penguins
| species | island | bill_length_mm | bill_depth_mm | flipper_length_mm | body_mass_g | sex | |
|---|---|---|---|---|---|---|---|
| 0 | Adelie | Torgersen | 39.1 | 18.7 | 181.0 | 3750.0 | Male |
| 1 | Adelie | Torgersen | 39.5 | 17.4 | 186.0 | 3800.0 | Female |
| 2 | Adelie | Torgersen | 40.3 | 18.0 | 195.0 | 3250.0 | Female |
| 4 | Adelie | Torgersen | 36.7 | 19.3 | 193.0 | 3450.0 | Female |
| 5 | Adelie | Torgersen | 39.3 | 20.6 | 190.0 | 3650.0 | Male |
| ... | ... | ... | ... | ... | ... | ... | ... |
| 338 | Gentoo | Biscoe | 47.2 | 13.7 | 214.0 | 4925.0 | Female |
| 340 | Gentoo | Biscoe | 46.8 | 14.3 | 215.0 | 4850.0 | Female |
| 341 | Gentoo | Biscoe | 50.4 | 15.7 | 222.0 | 5750.0 | Male |
| 342 | Gentoo | Biscoe | 45.2 | 14.8 | 212.0 | 5200.0 | Female |
| 343 | Gentoo | Biscoe | 49.9 | 16.1 | 213.0 | 5400.0 | Male |
333 rows × 7 columns
penguins.groupby('species')['body_mass_g'].transform(lambda ser: ser - ser.mean())
0 43.835616
1 93.835616
2 -456.164384
4 -256.164384
5 -56.164384
...
338 -167.436975
340 -242.436975
341 657.563025
342 107.563025
343 307.563025
Name: body_mass_g, Length: 333, dtype: float64
filter method¶filter method, which takes in a function.True.penguins
| species | island | bill_length_mm | bill_depth_mm | flipper_length_mm | body_mass_g | sex | |
|---|---|---|---|---|---|---|---|
| 0 | Adelie | Torgersen | 39.1 | 18.7 | 181.0 | 3750.0 | Male |
| 1 | Adelie | Torgersen | 39.5 | 17.4 | 186.0 | 3800.0 | Female |
| 2 | Adelie | Torgersen | 40.3 | 18.0 | 195.0 | 3250.0 | Female |
| 4 | Adelie | Torgersen | 36.7 | 19.3 | 193.0 | 3450.0 | Female |
| 5 | Adelie | Torgersen | 39.3 | 20.6 | 190.0 | 3650.0 | Male |
| ... | ... | ... | ... | ... | ... | ... | ... |
| 338 | Gentoo | Biscoe | 47.2 | 13.7 | 214.0 | 4925.0 | Female |
| 340 | Gentoo | Biscoe | 46.8 | 14.3 | 215.0 | 4850.0 | Female |
| 341 | Gentoo | Biscoe | 50.4 | 15.7 | 222.0 | 5750.0 | Male |
| 342 | Gentoo | Biscoe | 45.2 | 14.8 | 212.0 | 5200.0 | Female |
| 343 | Gentoo | Biscoe | 49.9 | 16.1 | 213.0 | 5400.0 | Male |
333 rows × 7 columns
penguins.groupby('species').filter(lambda df: df['bill_length_mm'].mean() > 39)
| species | island | bill_length_mm | bill_depth_mm | flipper_length_mm | body_mass_g | sex | |
|---|---|---|---|---|---|---|---|
| 152 | Chinstrap | Dream | 46.5 | 17.9 | 192.0 | 3500.0 | Female |
| 153 | Chinstrap | Dream | 50.0 | 19.5 | 196.0 | 3900.0 | Male |
| 154 | Chinstrap | Dream | 51.3 | 19.2 | 193.0 | 3650.0 | Male |
| 155 | Chinstrap | Dream | 45.4 | 18.7 | 188.0 | 3525.0 | Female |
| 156 | Chinstrap | Dream | 52.7 | 19.8 | 197.0 | 3725.0 | Male |
| ... | ... | ... | ... | ... | ... | ... | ... |
| 338 | Gentoo | Biscoe | 47.2 | 13.7 | 214.0 | 4925.0 | Female |
| 340 | Gentoo | Biscoe | 46.8 | 14.3 | 215.0 | 4850.0 | Female |
| 341 | Gentoo | Biscoe | 50.4 | 15.7 | 222.0 | 5750.0 | Male |
| 342 | Gentoo | Biscoe | 45.2 | 14.8 | 212.0 | 5200.0 | Female |
| 343 | Gentoo | Biscoe | 49.9 | 16.1 | 213.0 | 5400.0 | Male |
187 rows × 7 columns
No more Adelies!
apply method¶apply method is a generalization of aggregate, transform, and filter.apply.penguins.groupby('species').apply(lambda s: s * 2)
| species | island | bill_length_mm | bill_depth_mm | flipper_length_mm | body_mass_g | sex | |
|---|---|---|---|---|---|---|---|
| 0 | AdelieAdelie | TorgersenTorgersen | 78.2 | 37.4 | 362.0 | 7500.0 | MaleMale |
| 1 | AdelieAdelie | TorgersenTorgersen | 79.0 | 34.8 | 372.0 | 7600.0 | FemaleFemale |
| 2 | AdelieAdelie | TorgersenTorgersen | 80.6 | 36.0 | 390.0 | 6500.0 | FemaleFemale |
| 4 | AdelieAdelie | TorgersenTorgersen | 73.4 | 38.6 | 386.0 | 6900.0 | FemaleFemale |
| 5 | AdelieAdelie | TorgersenTorgersen | 78.6 | 41.2 | 380.0 | 7300.0 | MaleMale |
| ... | ... | ... | ... | ... | ... | ... | ... |
| 338 | GentooGentoo | BiscoeBiscoe | 94.4 | 27.4 | 428.0 | 9850.0 | FemaleFemale |
| 340 | GentooGentoo | BiscoeBiscoe | 93.6 | 28.6 | 430.0 | 9700.0 | FemaleFemale |
| 341 | GentooGentoo | BiscoeBiscoe | 100.8 | 31.4 | 444.0 | 11500.0 | MaleMale |
| 342 | GentooGentoo | BiscoeBiscoe | 90.4 | 29.6 | 424.0 | 10400.0 | FemaleFemale |
| 343 | GentooGentoo | BiscoeBiscoe | 99.8 | 32.2 | 426.0 | 10800.0 | MaleMale |
333 rows × 7 columns
penguins.groupby('species').apply(lambda s: s.mean().mean())
/var/folders/pd/w73mdrsj2836_7gp0brr2q7r0000gn/T/ipykernel_19120/2735591403.py:1: FutureWarning: Dropping of nuisance columns in DataFrame reductions (with 'numeric_only=None') is deprecated; in a future version this will raise TypeError. Select only valid columns before calling the reduction.
penguins.groupby('species').apply(lambda s: s.mean().mean())
species Adelie 988.359589 Chinstrap 999.041544 Gentoo 1343.059244 dtype: float64
For each species, find the island on which the heaviest penguin of that species lives.
# Why doesn't this work?
penguins.groupby('species').max()
| island | bill_length_mm | bill_depth_mm | flipper_length_mm | body_mass_g | sex | |
|---|---|---|---|---|---|---|
| species | ||||||
| Adelie | Torgersen | 46.0 | 21.5 | 210.0 | 4775.0 | Male |
| Chinstrap | Dream | 58.0 | 20.8 | 212.0 | 4800.0 | Male |
| Gentoo | Biscoe | 59.6 | 17.3 | 231.0 | 6300.0 | Male |
penguins.sort_values('body_mass_g', ascending=False).groupby('species').first()
| island | bill_length_mm | bill_depth_mm | flipper_length_mm | body_mass_g | sex | |
|---|---|---|---|---|---|---|
| species | ||||||
| Adelie | Biscoe | 43.2 | 19.0 | 197.0 | 4775.0 | Male |
| Chinstrap | Dream | 52.0 | 20.7 | 210.0 | 4800.0 | Male |
| Gentoo | Biscoe | 49.2 | 15.2 | 221.0 | 6300.0 | Male |
When we group with multiple columns, one group is created for every unique combination of elements in the specified columns.
double_group = penguins.groupby(['species', 'island'])
double_group
<pandas.core.groupby.generic.DataFrameGroupBy object at 0x7fae133cea90>
double_group.groups
{('Adelie', 'Biscoe'): [20, 21, 22, 23, 24, 25, 26, 27, 28, 29, 50, 51, 52, 53, 54, 55, 56, 57, 58, 59, 60, 61, 62, 63, 64, 65, 66, 67, 100, 101, 102, 103, 104, 105, 106, 107, 108, 109, 110, 111, 112, 113, 114, 115], ('Adelie', 'Dream'): [30, 31, 32, 33, 34, 35, 36, 37, 38, 39, 40, 41, 42, 43, 44, 45, 46, 48, 49, 84, 85, 86, 87, 88, 89, 90, 91, 92, 93, 94, 95, 96, 97, 98, 99, 132, 133, 134, 135, 136, 137, 138, 139, 140, 141, 142, 143, 144, 145, 146, 147, 148, 149, 150, 151], ('Adelie', 'Torgersen'): [0, 1, 2, 4, 5, 6, 7, 12, 13, 14, 15, 16, 17, 18, 19, 68, 69, 70, 71, 72, 73, 74, 75, 76, 77, 78, 79, 80, 81, 82, 83, 116, 117, 118, 119, 120, 121, 122, 123, 124, 125, 126, 127, 128, 129, 130, 131], ('Chinstrap', 'Dream'): [152, 153, 154, 155, 156, 157, 158, 159, 160, 161, 162, 163, 164, 165, 166, 167, 168, 169, 170, 171, 172, 173, 174, 175, 176, 177, 178, 179, 180, 181, 182, 183, 184, 185, 186, 187, 188, 189, 190, 191, 192, 193, 194, 195, 196, 197, 198, 199, 200, 201, 202, 203, 204, 205, 206, 207, 208, 209, 210, 211, 212, 213, 214, 215, 216, 217, 218, 219], ('Gentoo', 'Biscoe'): [220, 221, 222, 223, 224, 225, 226, 227, 228, 229, 230, 231, 232, 233, 234, 235, 236, 237, 238, 239, 240, 241, 242, 243, 244, 245, 247, 248, 249, 250, 251, 252, 253, 254, 255, 256, 257, 258, 259, 260, 261, 262, 263, 264, 265, 266, 267, 268, 269, 270, 271, 272, 273, 274, 275, 276, 277, 278, 279, 280, 281, 282, 283, 284, 285, 287, 288, 289, 290, 291, 292, 293, 294, 295, 296, 297, 298, 299, 300, 301, 302, 303, 304, 305, 306, 307, 308, 309, 310, 311, 312, 313, 314, 315, 316, 317, 318, 319, 320, 321, ...]}
for key, df in double_group:
display(df.head())
| species | island | bill_length_mm | bill_depth_mm | flipper_length_mm | body_mass_g | sex | |
|---|---|---|---|---|---|---|---|
| 20 | Adelie | Biscoe | 37.8 | 18.3 | 174.0 | 3400.0 | Female |
| 21 | Adelie | Biscoe | 37.7 | 18.7 | 180.0 | 3600.0 | Male |
| 22 | Adelie | Biscoe | 35.9 | 19.2 | 189.0 | 3800.0 | Female |
| 23 | Adelie | Biscoe | 38.2 | 18.1 | 185.0 | 3950.0 | Male |
| 24 | Adelie | Biscoe | 38.8 | 17.2 | 180.0 | 3800.0 | Male |
| species | island | bill_length_mm | bill_depth_mm | flipper_length_mm | body_mass_g | sex | |
|---|---|---|---|---|---|---|---|
| 30 | Adelie | Dream | 39.5 | 16.7 | 178.0 | 3250.0 | Female |
| 31 | Adelie | Dream | 37.2 | 18.1 | 178.0 | 3900.0 | Male |
| 32 | Adelie | Dream | 39.5 | 17.8 | 188.0 | 3300.0 | Female |
| 33 | Adelie | Dream | 40.9 | 18.9 | 184.0 | 3900.0 | Male |
| 34 | Adelie | Dream | 36.4 | 17.0 | 195.0 | 3325.0 | Female |
| species | island | bill_length_mm | bill_depth_mm | flipper_length_mm | body_mass_g | sex | |
|---|---|---|---|---|---|---|---|
| 0 | Adelie | Torgersen | 39.1 | 18.7 | 181.0 | 3750.0 | Male |
| 1 | Adelie | Torgersen | 39.5 | 17.4 | 186.0 | 3800.0 | Female |
| 2 | Adelie | Torgersen | 40.3 | 18.0 | 195.0 | 3250.0 | Female |
| 4 | Adelie | Torgersen | 36.7 | 19.3 | 193.0 | 3450.0 | Female |
| 5 | Adelie | Torgersen | 39.3 | 20.6 | 190.0 | 3650.0 | Male |
| species | island | bill_length_mm | bill_depth_mm | flipper_length_mm | body_mass_g | sex | |
|---|---|---|---|---|---|---|---|
| 152 | Chinstrap | Dream | 46.5 | 17.9 | 192.0 | 3500.0 | Female |
| 153 | Chinstrap | Dream | 50.0 | 19.5 | 196.0 | 3900.0 | Male |
| 154 | Chinstrap | Dream | 51.3 | 19.2 | 193.0 | 3650.0 | Male |
| 155 | Chinstrap | Dream | 45.4 | 18.7 | 188.0 | 3525.0 | Female |
| 156 | Chinstrap | Dream | 52.7 | 19.8 | 197.0 | 3725.0 | Male |
| species | island | bill_length_mm | bill_depth_mm | flipper_length_mm | body_mass_g | sex | |
|---|---|---|---|---|---|---|---|
| 220 | Gentoo | Biscoe | 46.1 | 13.2 | 211.0 | 4500.0 | Female |
| 221 | Gentoo | Biscoe | 50.0 | 16.3 | 230.0 | 5700.0 | Male |
| 222 | Gentoo | Biscoe | 48.7 | 14.1 | 210.0 | 4450.0 | Female |
| 223 | Gentoo | Biscoe | 50.0 | 15.2 | 218.0 | 5700.0 | Male |
| 224 | Gentoo | Biscoe | 47.6 | 14.5 | 215.0 | 5400.0 | Male |
penguins.groupby(['species', 'island']).mean()
| bill_length_mm | bill_depth_mm | flipper_length_mm | body_mass_g | ||
|---|---|---|---|---|---|
| species | island | ||||
| Adelie | Biscoe | 38.975000 | 18.370455 | 188.795455 | 3709.659091 |
| Dream | 38.520000 | 18.240000 | 189.927273 | 3701.363636 | |
| Torgersen | 39.038298 | 18.451064 | 191.531915 | 3708.510638 | |
| Chinstrap | Dream | 48.833824 | 18.420588 | 195.823529 | 3733.088235 |
| Gentoo | Biscoe | 47.568067 | 14.996639 | 217.235294 | 5092.436975 |
groupby method creates an index based on the specified columns.MultiIndex.MultiIndex, use reset_index or set as_index=False in groupby.weird = penguins.groupby(['species', 'island']).mean()
weird
| bill_length_mm | bill_depth_mm | flipper_length_mm | body_mass_g | ||
|---|---|---|---|---|---|
| species | island | ||||
| Adelie | Biscoe | 38.975000 | 18.370455 | 188.795455 | 3709.659091 |
| Dream | 38.520000 | 18.240000 | 189.927273 | 3701.363636 | |
| Torgersen | 39.038298 | 18.451064 | 191.531915 | 3708.510638 | |
| Chinstrap | Dream | 48.833824 | 18.420588 | 195.823529 | 3733.088235 |
| Gentoo | Biscoe | 47.568067 | 14.996639 | 217.235294 | 5092.436975 |
weird['body_mass_g']
species island
Adelie Biscoe 3709.659091
Dream 3701.363636
Torgersen 3708.510638
Chinstrap Dream 3733.088235
Gentoo Biscoe 5092.436975
Name: body_mass_g, dtype: float64
weird.loc['Adelie']
| bill_length_mm | bill_depth_mm | flipper_length_mm | body_mass_g | |
|---|---|---|---|---|
| island | ||||
| Biscoe | 38.975000 | 18.370455 | 188.795455 | 3709.659091 |
| Dream | 38.520000 | 18.240000 | 189.927273 | 3701.363636 |
| Torgersen | 39.038298 | 18.451064 | 191.531915 | 3708.510638 |
weird.loc[('Adelie', 'Torgersen')]
bill_length_mm 39.038298 bill_depth_mm 18.451064 flipper_length_mm 191.531915 body_mass_g 3708.510638 Name: (Adelie, Torgersen), dtype: float64
weird.reset_index()
| species | island | bill_length_mm | bill_depth_mm | flipper_length_mm | body_mass_g | |
|---|---|---|---|---|---|---|
| 0 | Adelie | Biscoe | 38.975000 | 18.370455 | 188.795455 | 3709.659091 |
| 1 | Adelie | Dream | 38.520000 | 18.240000 | 189.927273 | 3701.363636 |
| 2 | Adelie | Torgersen | 39.038298 | 18.451064 | 191.531915 | 3708.510638 |
| 3 | Chinstrap | Dream | 48.833824 | 18.420588 | 195.823529 | 3733.088235 |
| 4 | Gentoo | Biscoe | 47.568067 | 14.996639 | 217.235294 | 5092.436975 |
penguins.groupby(['species', 'island'], as_index=False).mean()
| species | island | bill_length_mm | bill_depth_mm | flipper_length_mm | body_mass_g | |
|---|---|---|---|---|---|---|
| 0 | Adelie | Biscoe | 38.975000 | 18.370455 | 188.795455 | 3709.659091 |
| 1 | Adelie | Dream | 38.520000 | 18.240000 | 189.927273 | 3701.363636 |
| 2 | Adelie | Torgersen | 39.038298 | 18.451064 | 191.531915 | 3708.510638 |
| 3 | Chinstrap | Dream | 48.833824 | 18.420588 | 195.823529 | 3733.088235 |
| 4 | Gentoo | Biscoe | 47.568067 | 14.996639 | 217.235294 | 5092.436975 |
To find the above information, we can group by both 'species' and 'island'.
penguins.groupby(['species', 'island'])['body_mass_g'].mean()
species island
Adelie Biscoe 3709.659091
Dream 3701.363636
Torgersen 3708.510638
Chinstrap Dream 3733.088235
Gentoo Biscoe 5092.436975
Name: body_mass_g, dtype: float64
But we can also create a pivot table.
penguins.pivot_table(index='species',
columns='island',
values='body_mass_g',
aggfunc='mean')
| island | Biscoe | Dream | Torgersen |
|---|---|---|---|
| species | |||
| Adelie | 3709.659091 | 3701.363636 | 3708.510638 |
| Chinstrap | NaN | 3733.088235 | NaN |
| Gentoo | 5092.436975 | NaN | NaN |
Note that the DataFrame above shows the same information as the Series above it, just in a different arrangement.
groupby method returns a DataFrameGroupBy method, which creates one group for every unique combination of values in the column(s) being grouped on.DataFrameGroupBy object, but we can also use transform, filter, or the more general apply methods. Each one of these methods acts on each group individually.pivot and pivot_table. Simpson's paradox. Combining DataFrames.