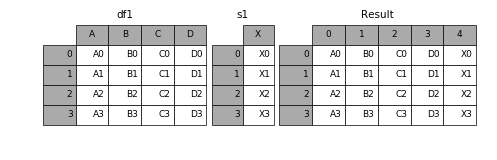
import pandas as pd
import numpy as np
import os
Good resource: pandas User Guide.
By default, pd.concat stacks DataFrames row-wise, i.e. on top of one another.
section_A = pd.DataFrame({
'Name': ['Annie', 'Billy', 'Sally', 'Tommy'],
'Midterm': [98, 82, 23, 45],
'Final': [88, 100, 99, 67]
})
section_A
| Name | Midterm | Final | |
|---|---|---|---|
| 0 | Annie | 98 | 88 |
| 1 | Billy | 82 | 100 |
| 2 | Sally | 23 | 99 |
| 3 | Tommy | 45 | 67 |
section_B = pd.DataFrame({
'Name': ['Junior', 'Rex', 'Flash'],
'Midterm': [70, 99, 81],
'Final': [42, 25, 90]
})
section_B
| Name | Midterm | Final | |
|---|---|---|---|
| 0 | Junior | 70 | 42 |
| 1 | Rex | 99 | 25 |
| 2 | Flash | 81 | 90 |
Let's use pd.concat on a list of the above two DataFrames.
pd.concat([section_A, section_B])
| Name | Midterm | Final | |
|---|---|---|---|
| 0 | Annie | 98 | 88 |
| 1 | Billy | 82 | 100 |
| 2 | Sally | 23 | 99 |
| 3 | Tommy | 45 | 67 |
| 0 | Junior | 70 | 42 |
| 1 | Rex | 99 | 25 |
| 2 | Flash | 81 | 90 |
pd.concat returns a copy; it does not modify any of the input DataFrames.pd.concat in a loop, as it has terrible time and space efficiency.total = pd.DataFrame()
for df in dataframes:
total = total.concat(df)
pd.concat(dataframes), where dataframes is a list of DataFrames.Suppose we have two DataFrames, exams and assignments, which both contain different attributes for the same individuals.
exams = section_A.copy()
exams
| Name | Midterm | Final | |
|---|---|---|---|
| 0 | Annie | 98 | 88 |
| 1 | Billy | 82 | 100 |
| 2 | Sally | 23 | 99 |
| 3 | Tommy | 45 | 67 |
assignments = exams[['Name']].assign(Homeworks=[99, 45, 23, 81],
Labs=[100, 100, 99, 100])
assignments
| Name | Homeworks | Labs | |
|---|---|---|---|
| 0 | Annie | 99 | 100 |
| 1 | Billy | 45 | 100 |
| 2 | Sally | 23 | 99 |
| 3 | Tommy | 81 | 100 |
If we try to combine these DataFrames with pd.concat, we don't quite get what we're looking for.
pd.concat([exams, assignments])
| Name | Midterm | Final | Homeworks | Labs | |
|---|---|---|---|---|---|
| 0 | Annie | 98.0 | 88.0 | NaN | NaN |
| 1 | Billy | 82.0 | 100.0 | NaN | NaN |
| 2 | Sally | 23.0 | 99.0 | NaN | NaN |
| 3 | Tommy | 45.0 | 67.0 | NaN | NaN |
| 0 | Annie | NaN | NaN | 99.0 | 100.0 |
| 1 | Billy | NaN | NaN | 45.0 | 100.0 |
| 2 | Sally | NaN | NaN | 23.0 | 99.0 |
| 3 | Tommy | NaN | NaN | 81.0 | 100.0 |
But that's where the axis argument becomes handy.
Remember, most pandas operations default to axis=0, but here we want to concatenate the columns of exams to the columns of assignments, so we should use axis=1.
pd.concat([exams, assignments], axis=1)
| Name | Midterm | Final | Name | Homeworks | Labs | |
|---|---|---|---|---|---|---|
| 0 | Annie | 98 | 88 | Annie | 99 | 100 |
| 1 | Billy | 82 | 100 | Billy | 45 | 100 |
| 2 | Sally | 23 | 99 | Sally | 23 | 99 |
| 3 | Tommy | 45 | 67 | Tommy | 81 | 100 |
Note that the 'Name' column appears twice!
pd.concat with axis=1.
Note that the call to pd.concat below works as expected, even though the orders of the names in exams_by_name and assignments_by_name are different.
# .loc[::-1] reverses the rows of the DataFrame
exams_by_name = exams.set_index('Name').loc[::-1]
exams_by_name
| Midterm | Final | |
|---|---|---|
| Name | ||
| Tommy | 45 | 67 |
| Sally | 23 | 99 |
| Billy | 82 | 100 |
| Annie | 98 | 88 |
assignments_by_name = assignments.set_index('Name')
assignments_by_name
| Homeworks | Labs | |
|---|---|---|
| Name | ||
| Annie | 99 | 100 |
| Billy | 45 | 100 |
| Sally | 23 | 99 |
| Tommy | 81 | 100 |
pd.concat([exams_by_name, assignments_by_name], axis=1)
| Midterm | Final | Homeworks | Labs | |
|---|---|---|---|---|
| Name | ||||
| Tommy | 45 | 67 | 81 | 100 |
| Sally | 23 | 99 | 23 | 99 |
| Billy | 82 | 100 | 45 | 100 |
| Annie | 98 | 88 | 99 | 100 |
Remember that pd.concat only looks at the index when combining rows, not at any other columns.
exams_reversed = exams.loc[::-1].reset_index(drop=True)
exams_reversed
| Name | Midterm | Final | |
|---|---|---|---|
| 0 | Tommy | 45 | 67 |
| 1 | Sally | 23 | 99 |
| 2 | Billy | 82 | 100 |
| 3 | Annie | 98 | 88 |
assignments
| Name | Homeworks | Labs | |
|---|---|---|---|
| 0 | Annie | 99 | 100 |
| 1 | Billy | 45 | 100 |
| 2 | Sally | 23 | 99 |
| 3 | Tommy | 81 | 100 |
pd.concat([exams_reversed, assignments], axis=1)
| Name | Midterm | Final | Name | Homeworks | Labs | |
|---|---|---|---|---|---|---|
| 0 | Tommy | 45 | 67 | Annie | 99 | 100 |
| 1 | Sally | 23 | 99 | Billy | 45 | 100 |
| 2 | Billy | 82 | 100 | Sally | 23 | 99 |
| 3 | Annie | 98 | 88 | Tommy | 81 | 100 |
If we concatenate two DataFrames that don't share row indexes, NaNs are added in the rows that aren't shared.
exams_extra = exams.copy()
exams_extra.loc[4] = ['Junior', 100, 100]
exams_extra
| Name | Midterm | Final | |
|---|---|---|---|
| 0 | Annie | 98 | 88 |
| 1 | Billy | 82 | 100 |
| 2 | Sally | 23 | 99 |
| 3 | Tommy | 45 | 67 |
| 4 | Junior | 100 | 100 |
assignments
| Name | Homeworks | Labs | |
|---|---|---|---|
| 0 | Annie | 99 | 100 |
| 1 | Billy | 45 | 100 |
| 2 | Sally | 23 | 99 |
| 3 | Tommy | 81 | 100 |
pd.concat([exams_extra, assignments], axis=1)
| Name | Midterm | Final | Name | Homeworks | Labs | |
|---|---|---|---|---|---|---|
| 0 | Annie | 98 | 88 | Annie | 99.0 | 100.0 |
| 1 | Billy | 82 | 100 | Billy | 45.0 | 100.0 |
| 2 | Sally | 23 | 99 | Sally | 23.0 | 99.0 |
| 3 | Tommy | 45 | 67 | Tommy | 81.0 | 100.0 |
| 4 | Junior | 100 | 100 | NaN | NaN | NaN |
pd.concat¶pd.concat "stitches" two or more DataFrames together.axis=0, the DataFrames are concatenated vertically based on column names (rows on top of rows).axis=1, the DataFrames are concatenated horizontally based on row indexes (columns next to columns).pd.concat with axis=1 combines DataFrames horizontally.merge method¶merge DataFrame method joins two tables by columns or indexes.pandas' word for "join".pandas function.Let's work with a small example.
temps = pd.DataFrame({
'City': ['San Diego', 'Toronto', 'Rome'],
'Temperature': [76, 28, 56]
})
temps
| City | Temperature | |
|---|---|---|
| 0 | San Diego | 76 |
| 1 | Toronto | 28 |
| 2 | Rome | 56 |
countries = pd.DataFrame({
'City': ['Toronto', 'Shanghai', 'San Diego'],
'Country': ['Canada', 'China', 'USA']
})
countries
| City | Country | |
|---|---|---|
| 0 | Toronto | Canada |
| 1 | Shanghai | China |
| 2 | San Diego | USA |
temps.merge(countries)
| City | Temperature | Country | |
|---|---|---|---|
| 0 | San Diego | 76 | USA |
| 1 | Toronto | 28 | Canada |
We didn't specify which columns to merge on, so it defaulted to 'City'.
'Rome' and 'Shanghai' do not appear in the merged DataFrame.'Rome' in the second DataFrame, and'Shanghai' in the first DataFrame.merge performs is an inner join, which keeps the intersection of the join keys.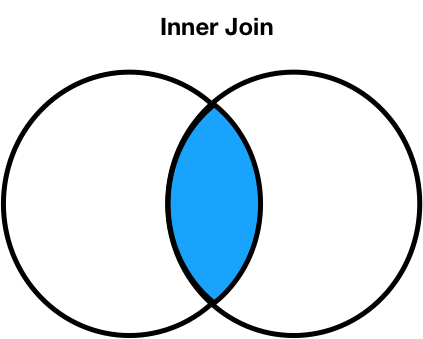
There are four types of joins.
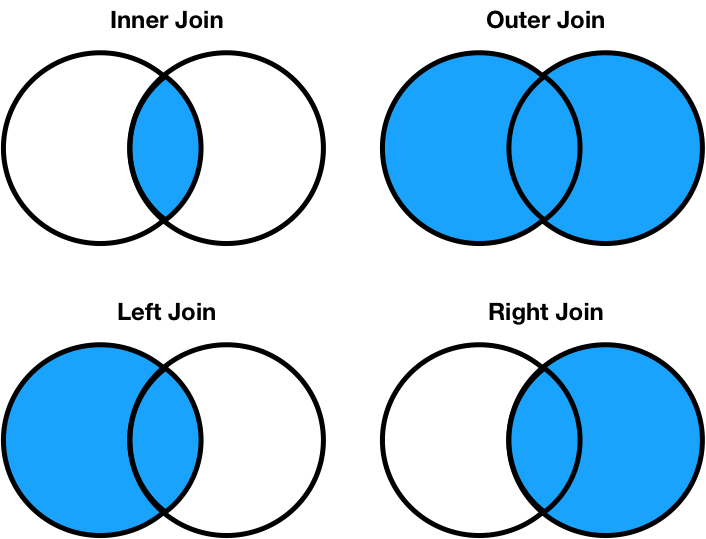
how argument (the default is how='inner')..merge, and the right DataFrame is the argument to merge.pd.merge function, in which the first argument is the left DataFrame and the second argument is the right DataFrame.temps
| City | Temperature | |
|---|---|---|
| 0 | San Diego | 76 |
| 1 | Toronto | 28 |
| 2 | Rome | 56 |
countries
| City | Country | |
|---|---|---|
| 0 | Toronto | Canada |
| 1 | Shanghai | China |
| 2 | San Diego | USA |
Let's try an outer join.
temps.merge(countries, how='outer')
| City | Temperature | Country | |
|---|---|---|---|
| 0 | San Diego | 76.0 | USA |
| 1 | Toronto | 28.0 | Canada |
| 2 | Rome | 56.0 | NaN |
| 3 | Shanghai | NaN | China |
# merge is also a pandas function
pd.merge(temps, countries, how='outer')
| City | Temperature | Country | |
|---|---|---|---|
| 0 | San Diego | 76.0 | USA |
| 1 | Toronto | 28.0 | Canada |
| 2 | Rome | 56.0 | NaN |
| 3 | Shanghai | NaN | China |
Note the NaNs in the rows for 'Rome' and 'Shanghai'.
Also note that an outer join is what pd.concat does by default, when there are no duplicated keys in either DataFrame.
pd.concat([temps.set_index('City'), countries.set_index('City')], axis=1)
| Temperature | Country | |
|---|---|---|
| City | ||
| San Diego | 76.0 | USA |
| Toronto | 28.0 | Canada |
| Rome | 56.0 | NaN |
| Shanghai | NaN | China |
Let's try left and right joins.
temps
| City | Temperature | |
|---|---|---|
| 0 | San Diego | 76 |
| 1 | Toronto | 28 |
| 2 | Rome | 56 |
countries
| City | Country | |
|---|---|---|
| 0 | Toronto | Canada |
| 1 | Shanghai | China |
| 2 | San Diego | USA |
First, a left join.
temps.merge(countries, how='left')
| City | Temperature | Country | |
|---|---|---|---|
| 0 | San Diego | 76 | USA |
| 1 | Toronto | 28 | Canada |
| 2 | Rome | 56 | NaN |
How about a right join?
temps.merge(countries, how='right')
| City | Temperature | Country | |
|---|---|---|---|
| 0 | Toronto | 28.0 | Canada |
| 1 | Shanghai | NaN | China |
| 2 | San Diego | 76.0 | USA |
Note that a.merge(b, how='left') is the same as b.merge(a, how='right'). The only difference is the order of the columns in the result.
countries.merge(temps, how='left')
| City | Country | Temperature | |
|---|---|---|---|
| 0 | Toronto | Canada | 28.0 |
| 1 | Shanghai | China | NaN |
| 2 | San Diego | USA | 76.0 |
pandas defaults to using the shared column(s) as join keys.on argument if the desired column(s) have the same names in both DataFrames.left_on or left_index argument AND the right_on or right_index argument.exams
| Name | Midterm | Final | |
|---|---|---|---|
| 0 | Annie | 98 | 88 |
| 1 | Billy | 82 | 100 |
| 2 | Sally | 23 | 99 |
| 3 | Tommy | 45 | 67 |
overall = pd.DataFrame({
'PID': ['A15253545', 'A10348245', 'A13349069', 'A18485824', 'A10094857'],
'Student': ['Billy', 'Sally', 'Annie', 'Larry', 'Johnny'],
'Final': [88, 64, 91, 45, 89]
})
overall
| PID | Student | Final | |
|---|---|---|---|
| 0 | A15253545 | Billy | 88 |
| 1 | A10348245 | Sally | 64 |
| 2 | A13349069 | Annie | 91 |
| 3 | A18485824 | Larry | 45 |
| 4 | A10094857 | Johnny | 89 |
This is not what we're looking for.
exams.merge(overall)
| Name | Midterm | Final | PID | Student | |
|---|---|---|---|---|---|
| 0 | Annie | 98 | 88 | A15253545 | Billy |
Instead, we need to tell pandas to look in the 'Name' column of exams and 'Student' column of overall.
exams.merge(overall, left_on='Name', right_on='Student')
| Name | Midterm | Final_x | PID | Student | Final_y | |
|---|---|---|---|---|---|---|
| 0 | Annie | 98 | 88 | A13349069 | Annie | 91 |
| 1 | Billy | 82 | 100 | A15253545 | Billy | 88 |
| 2 | Sally | 23 | 99 | A10348245 | Sally | 64 |
If there are shared column names in the two DataFrames you are merging that you are not using as join keys, by default '_x' and '_y' are appended to their names.
exams.merge(overall, left_on='Name', right_on='Student', suffixes=('_Exam', '_Overall'))
| Name | Midterm | Final_Exam | PID | Student | Final_Overall | |
|---|---|---|---|---|---|---|
| 0 | Annie | 98 | 88 | A13349069 | Annie | 91 |
| 1 | Billy | 82 | 100 | A15253545 | Billy | 88 |
| 2 | Sally | 23 | 99 | A10348245 | Sally | 64 |
If the desired join key is in the index, assign left_index or right_index to True.
exams
| Name | Midterm | Final | |
|---|---|---|---|
| 0 | Annie | 98 | 88 |
| 1 | Billy | 82 | 100 |
| 2 | Sally | 23 | 99 |
| 3 | Tommy | 45 | 67 |
overall_by_student = overall.set_index('Student')
overall_by_student
| PID | Final | |
|---|---|---|
| Student | ||
| Billy | A15253545 | 88 |
| Sally | A10348245 | 64 |
| Annie | A13349069 | 91 |
| Larry | A18485824 | 45 |
| Johnny | A10094857 | 89 |
exams.merge(overall_by_student, left_on='Name', right_index=True, suffixes=('_Exam', '_Overall'))
| Name | Midterm | Final_Exam | PID | Final_Overall | |
|---|---|---|---|---|---|
| 0 | Annie | 98 | 88 | A13349069 | 91 |
| 1 | Billy | 82 | 100 | A15253545 | 88 |
| 2 | Sally | 23 | 99 | A10348245 | 64 |
profs = pd.DataFrame(
[['Brad', 'UCB', 8],
['Janine', 'UCSD', 7],
['Marina', 'UIC', 6],
['Justin', 'OSU', 4],
['Aaron', 'UCB', 4],
['Soohyun', 'UCSD', 1],
['Suraj', 'UCB', 1]],
columns=['Name', 'School', 'Years']
)
profs
| Name | School | Years | |
|---|---|---|---|
| 0 | Brad | UCB | 8 |
| 1 | Janine | UCSD | 7 |
| 2 | Marina | UIC | 6 |
| 3 | Justin | OSU | 4 |
| 4 | Aaron | UCB | 4 |
| 5 | Soohyun | UCSD | 1 |
| 6 | Suraj | UCB | 1 |
schools = pd.DataFrame({
'Abr': ['UCSD', 'UCLA', 'UCB', 'UIC'],
'Full': ['University of California, San Diego', 'University of California, Los Angeles', 'University of California, Berkeley', 'University of Illinois Chicago']
})
schools
| Abr | Full | |
|---|---|---|
| 0 | UCSD | University of California, San Diego |
| 1 | UCLA | University of California, Los Angeles |
| 2 | UCB | University of California, Berkeley |
| 3 | UIC | University of Illinois Chicago |
Note that when merging profs and schools, the information from schools is duplicated ('University of California, San Diego' appears twice and 'University of California, Berkeley' appears three times).
profs.merge(schools, left_on='School', right_on='Abr', how='left')
| Name | School | Years | Abr | Full | |
|---|---|---|---|---|---|
| 0 | Brad | UCB | 8 | UCB | University of California, Berkeley |
| 1 | Janine | UCSD | 7 | UCSD | University of California, San Diego |
| 2 | Marina | UIC | 6 | UIC | University of Illinois Chicago |
| 3 | Justin | OSU | 4 | NaN | NaN |
| 4 | Aaron | UCB | 4 | UCB | University of California, Berkeley |
| 5 | Soohyun | UCSD | 1 | UCSD | University of California, San Diego |
| 6 | Suraj | UCB | 1 | UCB | University of California, Berkeley |
Many-to-many joins are joins where both DataFrames have duplicate values in the join key.
profs
| Name | School | Years | |
|---|---|---|---|
| 0 | Brad | UCB | 8 |
| 1 | Janine | UCSD | 7 |
| 2 | Marina | UIC | 6 |
| 3 | Justin | OSU | 4 |
| 4 | Aaron | UCB | 4 |
| 5 | Soohyun | UCSD | 1 |
| 6 | Suraj | UCB | 1 |
programs = pd.DataFrame({
'uni': ['UCSD', 'UCSD', 'UCSD', 'UCB', 'OSU', 'OSU'],
'dept': ['Math', 'HDSI', 'COGS', 'CS', 'Math', 'CS'],
'grad_students': [205, 54, 281, 439, 304, 193]
})
programs
| uni | dept | grad_students | |
|---|---|---|---|
| 0 | UCSD | Math | 205 |
| 1 | UCSD | HDSI | 54 |
| 2 | UCSD | COGS | 281 |
| 3 | UCB | CS | 439 |
| 4 | OSU | Math | 304 |
| 5 | OSU | CS | 193 |
Before running the following cell, try predicting the number of rows in the output.
profs.merge(programs, left_on='School', right_on='uni')
| Name | School | Years | uni | dept | grad_students | |
|---|---|---|---|---|---|---|
| 0 | Brad | UCB | 8 | UCB | CS | 439 |
| 1 | Aaron | UCB | 4 | UCB | CS | 439 |
| 2 | Suraj | UCB | 1 | UCB | CS | 439 |
| 3 | Janine | UCSD | 7 | UCSD | Math | 205 |
| 4 | Janine | UCSD | 7 | UCSD | HDSI | 54 |
| 5 | Janine | UCSD | 7 | UCSD | COGS | 281 |
| 6 | Soohyun | UCSD | 1 | UCSD | Math | 205 |
| 7 | Soohyun | UCSD | 1 | UCSD | HDSI | 54 |
| 8 | Soohyun | UCSD | 1 | UCSD | COGS | 281 |
| 9 | Justin | OSU | 4 | OSU | Math | 304 |
| 10 | Justin | OSU | 4 | OSU | CS | 193 |
merge stitched together every UCSD row in profs with every UCSD row in programs. profs and 3 in programs, there are $2 \cdot 3 = 6$ UCSD rows in the output. The same applies for all other schools.os.listdir(dirname) returns a list of the names of the files in the folder dirname.os.listdir('data')
['orders.csv', '.DS_Store', 'stops_2017.csv', 'military.csv', 'stops_2016.csv', 'race_codes.csv']
pathlib library allows you to do this.import pathlib
file_list = list(pathlib.Path().glob('data/stops*.csv')) # glob allows for pattern matching
file_list
[PosixPath('data/stops_2017.csv'), PosixPath('data/stops_2016.csv')]
! in front of a command in a Jupyter Notebook cell to run it on the command-line.!ls data/stops*.csv
data/stops_2016.csv data/stops_2017.csv
file_list
[PosixPath('data/stops_2017.csv'), PosixPath('data/stops_2016.csv')]
list_of_dfs = [pd.read_csv(file) for file in file_list]
for df in list_of_dfs:
display(df.head())
| stop_id | stop_cause | service_area | subject_race | subject_sex | subject_age | timestamp | stop_date | stop_time | sd_resident | arrested | searched | obtained_consent | contraband_found | property_seized | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 1444799 | Moving Violation | 120 | I | M | 37 | 2017-01-01 00:03:00 | 2017-01-01 | 00:03:00 | N | N | N | NaN | NaN | NaN |
| 1 | 1444821 | Equipment Violation | 520 | W | M | 22 | 2017-01-01 00:25:00 | 2017-01-01 | 00:25:00 | N | N | N | NaN | NaN | NaN |
| 2 | 1447102 | Moving Violation | 520 | W | M | 29 | 2017-01-01 01:45:00 | 2017-01-01 | 01:45:00 | N | N | N | NaN | NaN | NaN |
| 3 | 1444801 | Equipment Violation | 720 | H | F | 61 | 2017-01-01 03:10:00 | 2017-01-01 | 03:10:00 | N | N | N | NaN | NaN | NaN |
| 4 | 1444802 | Equipment Violation | 120 | H | M | 24 | 2017-01-01 03:30:00 | 2017-01-01 | 03:30:00 | Y | N | N | NaN | NaN | NaN |
| stop_id | stop_cause | service_area | subject_race | subject_sex | subject_age | timestamp | stop_date | stop_time | sd_resident | arrested | searched | obtained_consent | contraband_found | property_seized | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 1308198 | Equipment Violation | 530 | W | M | 28 | 2016-01-01 00:06:00 | 2016-01-01 | 0:06 | Y | N | N | N | N | N |
| 1 | 1308172 | Moving Violation | 520 | B | M | 25 | 2016-01-01 00:10:00 | 2016-01-01 | 0:10 | N | N | N | NaN | NaN | NaN |
| 2 | 1308171 | Moving Violation | 110 | H | F | 31 | 2016-01-01 00:14:00 | 2016-01-01 | 0:14 | NaN | NaN | NaN | NaN | NaN | NaN |
| 3 | 1308170 | Moving Violation | Unknown | W | F | 29 | 2016-01-01 00:16:00 | 2016-01-01 | 0:16 | N | N | N | NaN | NaN | NaN |
| 4 | 1308197 | Moving Violation | 230 | W | M | 52 | 2016-01-01 00:30:00 | 2016-01-01 | 0:30 | N | N | N | NaN | NaN | NaN |
We need to concatenate these two DataFrames vertically.
stops = pd.concat(list_of_dfs, ignore_index=True)
stops.head()
| stop_id | stop_cause | service_area | subject_race | subject_sex | subject_age | timestamp | stop_date | stop_time | sd_resident | arrested | searched | obtained_consent | contraband_found | property_seized | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 1444799 | Moving Violation | 120 | I | M | 37 | 2017-01-01 00:03:00 | 2017-01-01 | 00:03:00 | N | N | N | NaN | NaN | NaN |
| 1 | 1444821 | Equipment Violation | 520 | W | M | 22 | 2017-01-01 00:25:00 | 2017-01-01 | 00:25:00 | N | N | N | NaN | NaN | NaN |
| 2 | 1447102 | Moving Violation | 520 | W | M | 29 | 2017-01-01 01:45:00 | 2017-01-01 | 01:45:00 | N | N | N | NaN | NaN | NaN |
| 3 | 1444801 | Equipment Violation | 720 | H | F | 61 | 2017-01-01 03:10:00 | 2017-01-01 | 03:10:00 | N | N | N | NaN | NaN | NaN |
| 4 | 1444802 | Equipment Violation | 120 | H | M | 24 | 2017-01-01 03:30:00 | 2017-01-01 | 03:30:00 | Y | N | N | NaN | NaN | NaN |
len(stops)
206413
Right now, 'subject_race' is stored as a single character. What does 'I' mean? 'H'?
stops.head()
| stop_id | stop_cause | service_area | subject_race | subject_sex | subject_age | timestamp | stop_date | stop_time | sd_resident | arrested | searched | obtained_consent | contraband_found | property_seized | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 1444799 | Moving Violation | 120 | I | M | 37 | 2017-01-01 00:03:00 | 2017-01-01 | 00:03:00 | N | N | N | NaN | NaN | NaN |
| 1 | 1444821 | Equipment Violation | 520 | W | M | 22 | 2017-01-01 00:25:00 | 2017-01-01 | 00:25:00 | N | N | N | NaN | NaN | NaN |
| 2 | 1447102 | Moving Violation | 520 | W | M | 29 | 2017-01-01 01:45:00 | 2017-01-01 | 01:45:00 | N | N | N | NaN | NaN | NaN |
| 3 | 1444801 | Equipment Violation | 720 | H | F | 61 | 2017-01-01 03:10:00 | 2017-01-01 | 03:10:00 | N | N | N | NaN | NaN | NaN |
| 4 | 1444802 | Equipment Violation | 120 | H | M | 24 | 2017-01-01 03:30:00 | 2017-01-01 | 03:30:00 | Y | N | N | NaN | NaN | NaN |
stops['subject_race'].value_counts(normalize=True)
W 0.411358 H 0.309249 B 0.112620 O 0.086896 A 0.046397 F 0.011923 V 0.005768 C 0.004899 I 0.004123 X 0.001446 K 0.001222 P 0.000990 J 0.000912 Z 0.000558 D 0.000543 L 0.000461 S 0.000349 G 0.000189 U 0.000097 Name: subject_race, dtype: float64
Fortunately, we have access to another dataset that describes each of the race codes.
races = pd.read_csv('data/race_codes.csv')
races
| Race Code | Description | |
|---|---|---|
| 0 | A | OTHER ASIAN |
| 1 | B | BLACK |
| 2 | C | CHINESE |
| 3 | D | CAMBODIAN |
| 4 | F | FILIPINO |
| 5 | G | GUAMANIAN |
| 6 | H | HISPANIC |
| 7 | I | INDIAN |
| 8 | J | JAPANESE |
| 9 | K | KOREAN |
| 10 | L | LAOTIAN |
| 11 | O | OTHER |
| 12 | P | PACIFIC ISLANDER |
| 13 | S | SAMOAN |
| 14 | U | HAWAIIAN |
| 15 | V | VIETNAMESE |
| 16 | W | WHITE |
| 17 | Z | ASIAN INDIAN |
Let's join the distribution of races with the DataFrame of race codes.
Question: Is this a one-to-one join?
race_percentages = stops['subject_race'].value_counts(normalize=True).rename('Proportion').to_frame()
race_percentages.merge(races, left_index=True, right_on='Race Code')
| Proportion | Race Code | Description | |
|---|---|---|---|
| 16 | 0.411358 | W | WHITE |
| 6 | 0.309249 | H | HISPANIC |
| 1 | 0.112620 | B | BLACK |
| 11 | 0.086896 | O | OTHER |
| 0 | 0.046397 | A | OTHER ASIAN |
| 4 | 0.011923 | F | FILIPINO |
| 15 | 0.005768 | V | VIETNAMESE |
| 2 | 0.004899 | C | CHINESE |
| 7 | 0.004123 | I | INDIAN |
| 9 | 0.001222 | K | KOREAN |
| 12 | 0.000990 | P | PACIFIC ISLANDER |
| 8 | 0.000912 | J | JAPANESE |
| 17 | 0.000558 | Z | ASIAN INDIAN |
| 3 | 0.000543 | D | CAMBODIAN |
| 10 | 0.000461 | L | LAOTIAN |
| 13 | 0.000349 | S | SAMOAN |
| 5 | 0.000189 | G | GUAMANIAN |
| 14 | 0.000097 | U | HAWAIIAN |
The level of granularity of the races in our data right now seems inconsistent. For instance, 'WHITE' and 'BLACK' are much more broad than 'FILIPINO', 'JAPANESE', and 'GUAMANIAN'.
Let's try and adjust our race data so that we have a consistent level of granularity. Here's what we want to create:
| Race Code | Description | Race_Category | |
|---|---|---|---|
| 0 | A | OTHER ASIAN | Asian |
| 1 | B | BLACK | Black |
| 2 | C | CHINESE | Asian |
| 3 | D | CAMBODIAN | Asian |
| 4 | F | FILIPINO | Asian |
We can do this by manually defining a mapping between race codes and desired categories.
race_dict = {'A':'Asian',
'B':'Black',
'C':'Asian',
'D':'Asian',
'F':'Asian',
'G':'Asian',
'H':'Hispanic',
'I':'Native American',
'J':'Asian',
'K':'Asian',
'L':'Asian',
'O':'Other',
'P':'Asian',
'S':'Asian',
'U':'Hawaiian',
'V':'Asian',
'W':'White',
'Z':'Asian'
}
There are two ways to replace all of the 'Race Code's in races with the above categories:
replace method.races.Joining requires sorting, where as replacing does not. Let's go with the first option.
races['Race_Category'] = races['Race Code'].replace(race_dict)
races
| Race Code | Description | Race_Category | |
|---|---|---|---|
| 0 | A | OTHER ASIAN | Asian |
| 1 | B | BLACK | Black |
| 2 | C | CHINESE | Asian |
| 3 | D | CAMBODIAN | Asian |
| 4 | F | FILIPINO | Asian |
| 5 | G | GUAMANIAN | Asian |
| 6 | H | HISPANIC | Hispanic |
| 7 | I | INDIAN | Native American |
| 8 | J | JAPANESE | Asian |
| 9 | K | KOREAN | Asian |
| 10 | L | LAOTIAN | Asian |
| 11 | O | OTHER | Other |
| 12 | P | PACIFIC ISLANDER | Asian |
| 13 | S | SAMOAN | Asian |
| 14 | U | HAWAIIAN | Hawaiian |
| 15 | V | VIETNAMESE | Asian |
| 16 | W | WHITE | White |
| 17 | Z | ASIAN INDIAN | Asian |
Now, we need to join stops with races. An important question is, what type of join should we use (inner, outer, left, right)?
stops.head()
| stop_id | stop_cause | service_area | subject_race | subject_sex | subject_age | timestamp | stop_date | stop_time | sd_resident | arrested | searched | obtained_consent | contraband_found | property_seized | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 1444799 | Moving Violation | 120 | I | M | 37 | 2017-01-01 00:03:00 | 2017-01-01 | 00:03:00 | N | N | N | NaN | NaN | NaN |
| 1 | 1444821 | Equipment Violation | 520 | W | M | 22 | 2017-01-01 00:25:00 | 2017-01-01 | 00:25:00 | N | N | N | NaN | NaN | NaN |
| 2 | 1447102 | Moving Violation | 520 | W | M | 29 | 2017-01-01 01:45:00 | 2017-01-01 | 01:45:00 | N | N | N | NaN | NaN | NaN |
| 3 | 1444801 | Equipment Violation | 720 | H | F | 61 | 2017-01-01 03:10:00 | 2017-01-01 | 03:10:00 | N | N | N | NaN | NaN | NaN |
| 4 | 1444802 | Equipment Violation | 120 | H | M | 24 | 2017-01-01 03:30:00 | 2017-01-01 | 03:30:00 | Y | N | N | NaN | NaN | NaN |
stops['subject_race'].isna().mean()
0.0012547659304404277
So that we don't discard the individuals whose races we don't have, we will use a left join.
stops_merged = stops.merge(races, left_on='subject_race', right_on='Race Code', how='left')
stops_merged.head()
| stop_id | stop_cause | service_area | subject_race | subject_sex | subject_age | timestamp | stop_date | stop_time | sd_resident | arrested | searched | obtained_consent | contraband_found | property_seized | Race Code | Description | Race_Category | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 1444799 | Moving Violation | 120 | I | M | 37 | 2017-01-01 00:03:00 | 2017-01-01 | 00:03:00 | N | N | N | NaN | NaN | NaN | I | INDIAN | Native American |
| 1 | 1444821 | Equipment Violation | 520 | W | M | 22 | 2017-01-01 00:25:00 | 2017-01-01 | 00:25:00 | N | N | N | NaN | NaN | NaN | W | WHITE | White |
| 2 | 1447102 | Moving Violation | 520 | W | M | 29 | 2017-01-01 01:45:00 | 2017-01-01 | 01:45:00 | N | N | N | NaN | NaN | NaN | W | WHITE | White |
| 3 | 1444801 | Equipment Violation | 720 | H | F | 61 | 2017-01-01 03:10:00 | 2017-01-01 | 03:10:00 | N | N | N | NaN | NaN | NaN | H | HISPANIC | Hispanic |
| 4 | 1444802 | Equipment Violation | 120 | H | M | 24 | 2017-01-01 03:30:00 | 2017-01-01 | 03:30:00 | Y | N | N | NaN | NaN | NaN | H | HISPANIC | Hispanic |
Now we can compute a more meaningful distribution of races.
dist = stops_merged['Race_Category'].value_counts(normalize=True)
dist
White 0.411953 Hispanic 0.309697 Black 0.112783 Other 0.087022 Asian 0.074319 Native American 0.004129 Hawaiian 0.000097 Name: Race_Category, dtype: float64
dist.plot(kind='bar', figsize=(10, 5));
That's a bit more helpful.
.csv file per day for 1 year.pd.concat the DataFrames together.pandas.When working with time data, you will see two different kinds of "times":
datetime module¶Python has an in-built datetime module, which contains datetime and timedelta types. These are much more convenient to deal with than strings that contain times.
import datetime
datetime.datetime.now()
datetime.datetime(2022, 4, 15, 13, 3, 11, 143648)
datetime.datetime.now() + datetime.timedelta(days=3, hours=5)
datetime.datetime(2022, 4, 18, 18, 3, 11, 147079)
Recall, Unix timestamps count the number of seconds since January 1st, 1970.
datetime.datetime.now().timestamp()
1650052991.150548
pandas¶pd.Timestamp is the pandas equivalent of datetime.pd.to_datetime converts strings to pd.Timestamp objects.pd.Timestamp(year=1998, month=11, day=26)
Timestamp('1998-11-26 00:00:00')
final_start = pd.to_datetime('June 4th, 2022, 11:30AM')
final_start
Timestamp('2022-06-04 11:30:00')
final_finish = pd.to_datetime('June 4th, 2022, 2:30PM')
final_finish
Timestamp('2022-06-04 14:30:00')
Timestamps have time-related attributes, e.g. dayofweek, hour, min, sec.
final_finish.dayofweek
5
final_finish.year
2022
Subtracting timestamps yields pd.Timedelta objects.
final_finish - final_start
Timedelta('0 days 03:00:00')
pd.to_datetime, pandas stores them as yet another type:
np.datetime64.pd.Timestamp, but optimized for memory and speed efficiency.pd.Timestamp back.times = pd.DataFrame({'finish': pd.to_datetime(['Sun, Jan 01, 1989',
'2022-04-15T11:00',
'1/1/1970'])})
times
| finish | |
|---|---|
| 0 | 1989-01-01 00:00:00 |
| 1 | 2022-04-15 11:00:00 |
| 2 | 1970-01-01 00:00:00 |
times.info()
<class 'pandas.core.frame.DataFrame'> RangeIndex: 3 entries, 0 to 2 Data columns (total 1 columns): # Column Non-Null Count Dtype --- ------ -------------- ----- 0 finish 3 non-null datetime64[ns] dtypes: datetime64[ns](1) memory usage: 152.0 bytes
times.iloc[0, 0]
Timestamp('1989-01-01 00:00:00')
times.sort_values('finish')
| finish | |
|---|---|
| 2 | 1970-01-01 00:00:00 |
| 0 | 1989-01-01 00:00:00 |
| 1 | 2022-04-15 11:00:00 |
Below, we have the Final Exam starting and ending times for two sections of a course.
times_A = pd.DataFrame({
'Name': ['Annie', 'Billy', 'Sally', 'Tommy'],
'start_exam': ['15:00', '15:02', '15:01', '15:00'],
'finish_exam': ['16:00', '17:58', '17:05', '16:55']
})
times_B = pd.DataFrame({
'Name': ['Junior', 'Rex', 'Flash'],
'start_exam': ['18:00', '18:06', '19:07'],
'finish_exam': ['20:00', '20:50', '20:59']
})
display(times_A)
display(times_B)
| Name | start_exam | finish_exam | |
|---|---|---|---|
| 0 | Annie | 15:00 | 16:00 |
| 1 | Billy | 15:02 | 17:58 |
| 2 | Sally | 15:01 | 17:05 |
| 3 | Tommy | 15:00 | 16:55 |
| Name | start_exam | finish_exam | |
|---|---|---|---|
| 0 | Junior | 18:00 | 20:00 |
| 1 | Rex | 18:06 | 20:50 |
| 2 | Flash | 19:07 | 20:59 |
Question: Who finished the exam the fastest amongst all students in the course?
Approach:
pd.Timestamp.'finish_exam' and 'start_exam'.# Step 1
both_versions = pd.concat([times_A, times_B])
both_versions
| Name | start_exam | finish_exam | |
|---|---|---|---|
| 0 | Annie | 15:00 | 16:00 |
| 1 | Billy | 15:02 | 17:58 |
| 2 | Sally | 15:01 | 17:05 |
| 3 | Tommy | 15:00 | 16:55 |
| 0 | Junior | 18:00 | 20:00 |
| 1 | Rex | 18:06 | 20:50 |
| 2 | Flash | 19:07 | 20:59 |
# Step 2
both_versions = both_versions.assign(
start_exam=pd.to_datetime(both_versions['start_exam']),
finish_exam=pd.to_datetime(both_versions['finish_exam'])
)
both_versions.info()
<class 'pandas.core.frame.DataFrame'> Int64Index: 7 entries, 0 to 2 Data columns (total 3 columns): # Column Non-Null Count Dtype --- ------ -------------- ----- 0 Name 7 non-null object 1 start_exam 7 non-null datetime64[ns] 2 finish_exam 7 non-null datetime64[ns] dtypes: datetime64[ns](2), object(1) memory usage: 224.0+ bytes
# Step 3
both_versions = both_versions.assign(
elapsed=both_versions['finish_exam'] - both_versions['start_exam']
)
both_versions
| Name | start_exam | finish_exam | elapsed | |
|---|---|---|---|---|
| 0 | Annie | 2022-04-15 15:00:00 | 2022-04-15 16:00:00 | 0 days 01:00:00 |
| 1 | Billy | 2022-04-15 15:02:00 | 2022-04-15 17:58:00 | 0 days 02:56:00 |
| 2 | Sally | 2022-04-15 15:01:00 | 2022-04-15 17:05:00 | 0 days 02:04:00 |
| 3 | Tommy | 2022-04-15 15:00:00 | 2022-04-15 16:55:00 | 0 days 01:55:00 |
| 0 | Junior | 2022-04-15 18:00:00 | 2022-04-15 20:00:00 | 0 days 02:00:00 |
| 1 | Rex | 2022-04-15 18:06:00 | 2022-04-15 20:50:00 | 0 days 02:44:00 |
| 2 | Flash | 2022-04-15 19:07:00 | 2022-04-15 20:59:00 | 0 days 01:52:00 |
# Steps 4 and 5
both_versions.sort_values('elapsed').iloc[0].loc['Name']
'Annie'
pd.concat "stitches" two or more DataFrames together, either vertically or horizontally.merge DataFrame method joins two DataFrames together based on a shared column, called a join key. There are four types of joins:NaNs.pandas are stored using pd.Timestamp and pd.Timedelta objects.