import numpy as np
import pandas as pd
import pathlib
import os
import matplotlib.pyplot as plt
import seaborn as sns
plt.style.use('seaborn-white')
plt.rc('figure', dpi=100, figsize=(10, 5))
plt.rc('font', size=12)


Credits to Nicole Brye

Great reading: jwilber.me/permutationtest.
So far, our hypothesis tests have assessed a model given a single random sample.
Often have two random samples we wish to compare.
Let's start by loading in data.
# Kaiser dataset, 70s
baby_fp = os.path.join('data', 'baby.csv')
baby = pd.read_csv(baby_fp)
baby.head()
| Birth Weight | Gestational Days | Maternal Age | Maternal Height | Maternal Pregnancy Weight | Maternal Smoker | |
|---|---|---|---|---|---|---|
| 0 | 120 | 284 | 27 | 62 | 100 | False |
| 1 | 113 | 282 | 33 | 64 | 135 | False |
| 2 | 128 | 279 | 28 | 64 | 115 | True |
| 3 | 108 | 282 | 23 | 67 | 125 | True |
| 4 | 136 | 286 | 25 | 62 | 93 | False |
Only the 'Birth Weight' and 'Maternal Smoker' columns are relevant.
smoking_and_birthweight = baby[['Maternal Smoker', 'Birth Weight']]
smoking_and_birthweight.head()
| Maternal Smoker | Birth Weight | |
|---|---|---|
| 0 | False | 120 |
| 1 | False | 113 |
| 2 | True | 128 |
| 3 | True | 108 |
| 4 | False | 136 |
How many babies are in each group?
smoking_and_birthweight.groupby('Maternal Smoker').count()
| Birth Weight | |
|---|---|
| Maternal Smoker | |
| False | 715 |
| True | 459 |
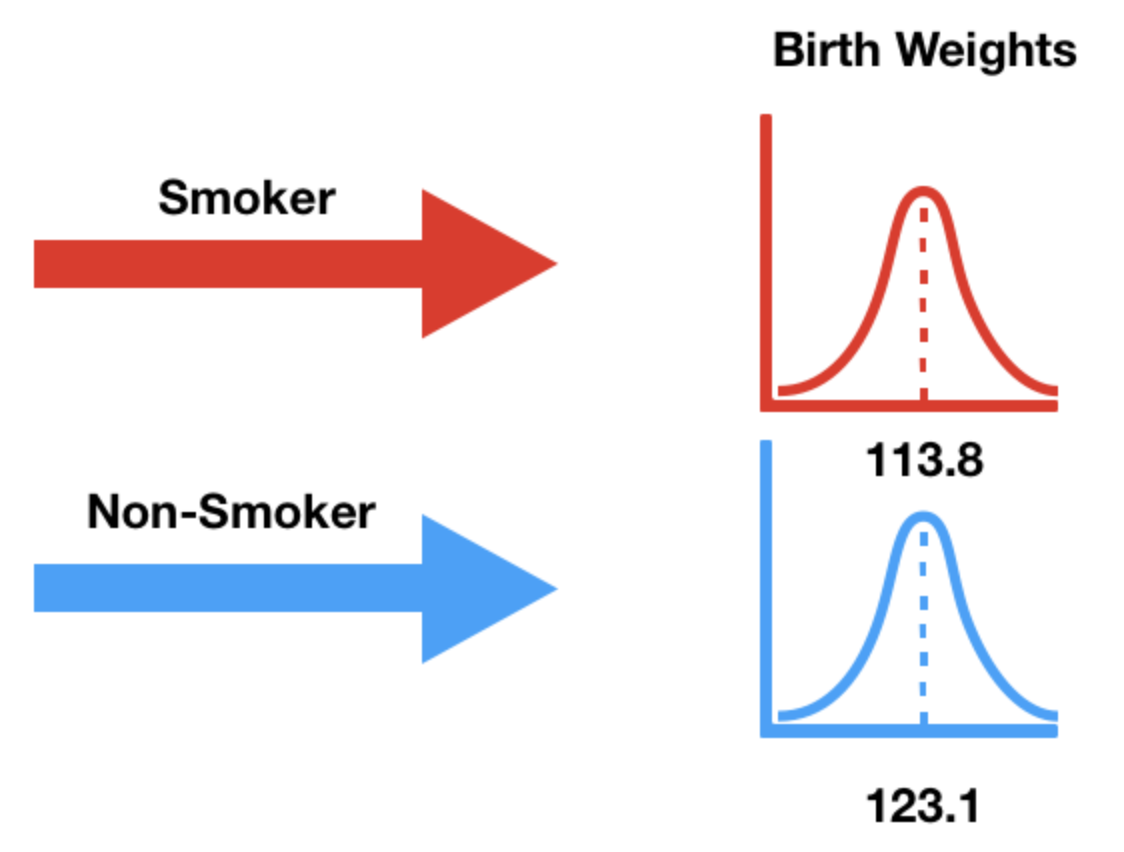
What is the average birth weight within each group?
smoking_and_birthweight.groupby('Maternal Smoker').mean()
| Birth Weight | |
|---|---|
| Maternal Smoker | |
| False | 123.085315 |
| True | 113.819172 |
Note that 16 ounces are in 1 pound, so the above weights are ~7-8 pounds.
title = "Birth Weight by Mother's Smoking Status"
(
smoking_and_birthweight
.groupby('Maternal Smoker')['Birth Weight']
.plot(kind='hist', density=True, legend=True,
ec='w', bins=np.arange(50, 200, 5), alpha=0.75,
title=title)
);
(
smoking_and_birthweight
.groupby('Maternal Smoker')['Birth Weight']
.plot(kind='kde', legend=True,
title=title)
);
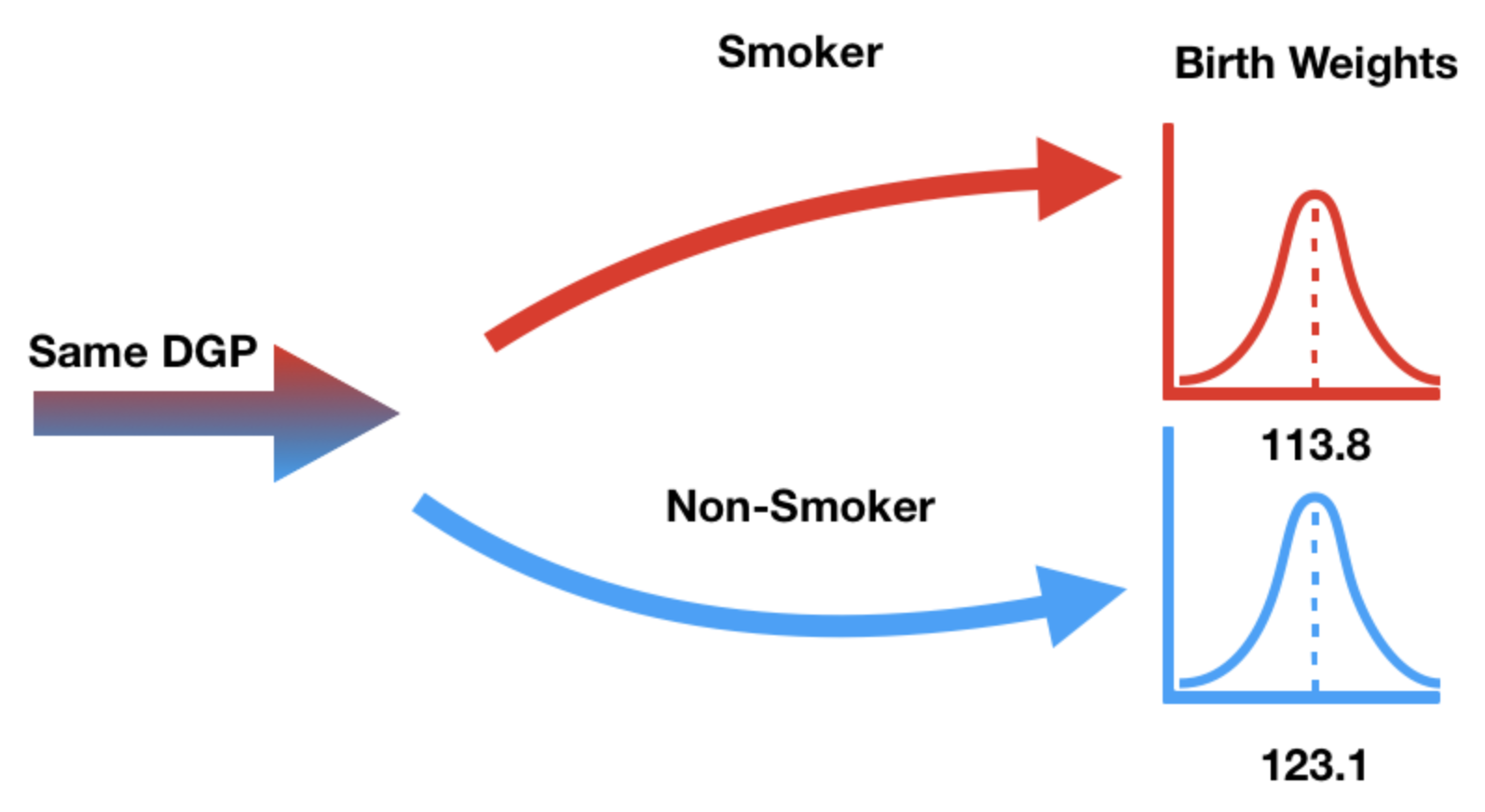
We need a test statistic that can measure how different two numerical distributions are.
(
smoking_and_birthweight
.groupby('Maternal Smoker')['Birth Weight']
.plot(kind='kde', legend=True,
title=title,
figsize=(4, 3))
);
Easiest solution: Difference in group means.
To compute the difference between the mean birth weight of babies born to smokers and the mean birth weight of babies born to non-smokers, we can use groupby.
smoking_and_birthweight.head()
| Maternal Smoker | Birth Weight | |
|---|---|---|
| 0 | False | 120 |
| 1 | False | 113 |
| 2 | True | 128 |
| 3 | True | 108 |
| 4 | False | 136 |
means_table = smoking_and_birthweight.groupby('Maternal Smoker').mean()
means_table
| Birth Weight | |
|---|---|
| Maternal Smoker | |
| False | 123.085315 |
| True | 113.819172 |
means_table.loc[True, 'Birth Weight'] - means_table.loc[False, 'Birth Weight']
-9.266142572024918
Note that we arbitrarily chose to compute the "smoking" mean minus the "non-smoking" mean. We could have chosen the other direction, too.
Insteading of using .loc and manually subtracting, there is another method we can use to find the difference in group means – the diff Series/DataFrame method.
s = pd.Series([1, 2, 5, 9, 15])
s
0 1 1 2 2 5 3 9 4 15 dtype: int64
s.diff()
0 NaN 1 1.0 2 3.0 3 4.0 4 6.0 dtype: float64
means_table
| Birth Weight | |
|---|---|
| Maternal Smoker | |
| False | 123.085315 |
| True | 113.819172 |
means_table.diff()
| Birth Weight | |
|---|---|
| Maternal Smoker | |
| False | NaN |
| True | -9.266143 |
observed_difference = means_table.diff().iloc[-1, 0]
observed_difference
-9.266142572024918
'Maternal Smoker' – has no effect on the birth weight.True or False to each baby.smoking_and_birthweight.head()
| Maternal Smoker | Birth Weight | |
|---|---|---|
| 0 | False | 120 |
| 1 | False | 113 |
| 2 | True | 128 |
| 3 | True | 108 |
| 4 | False | 136 |
True or False, while keeping the same number of Trues and Falses as we started with.'Maternal Smoker' column.sample method returns a random sample of rows in the DataFrame.n=df.shape[0] or frac=1.sample samples without replacement, which is what we want.smoking_and_birthweight.sample(frac=1)
| Maternal Smoker | Birth Weight | |
|---|---|---|
| 803 | False | 116 |
| 429 | False | 112 |
| 557 | False | 170 |
| 351 | True | 135 |
| 1053 | False | 141 |
| ... | ... | ... |
| 214 | False | 125 |
| 1013 | True | 103 |
| 254 | True | 123 |
| 459 | False | 130 |
| 532 | True | 128 |
1174 rows × 2 columns
df.sample, both columns are shuffled together.False, they will still be assigned to False in the shuffled version.True or False.smoking_and_birthweight.head()
| Maternal Smoker | Birth Weight | |
|---|---|---|
| 0 | False | 120 |
| 1 | False | 113 |
| 2 | True | 128 |
| 3 | True | 108 |
| 4 | False | 136 |
Remember, it doesn't matter which column we shuffle! Here, we'll shuffle birth weights.
shuffled_weights = (
smoking_and_birthweight['Birth Weight']
.sample(frac=1)
.reset_index(drop=True) # Question: What will happen if we do not reset the index?
)
shuffled_weights.head()
0 86 1 130 2 123 3 115 4 119 Name: Birth Weight, dtype: int64
original_and_shuffled = (
smoking_and_birthweight
.assign(**{'Shuffled Birth Weight': shuffled_weights})
)
original_and_shuffled.head(10)
| Maternal Smoker | Birth Weight | Shuffled Birth Weight | |
|---|---|---|---|
| 0 | False | 120 | 86 |
| 1 | False | 113 | 130 |
| 2 | True | 128 | 123 |
| 3 | True | 108 | 115 |
| 4 | False | 136 | 119 |
| 5 | False | 138 | 109 |
| 6 | False | 132 | 137 |
| 7 | False | 120 | 87 |
| 8 | True | 143 | 103 |
| 9 | False | 140 | 108 |
For details on how ** works, see this article.
One benefit of shuffling 'Birth Weight' (instead of 'Maternal Smoker') is that grouping by 'Maternal Smoker' allows us to see all of the following information with a single call to groupby.
original_and_shuffled.groupby('Maternal Smoker').mean()
| Birth Weight | Shuffled Birth Weight | |
|---|---|---|
| Maternal Smoker | ||
| False | 123.085315 | 119.995804 |
| True | 113.819172 | 118.631808 |
fig, axes = plt.subplots(1, 2, figsize=(15, 5))
title = 'Birth Weights by Maternal Smoker, SHUFFLED'
original_and_shuffled.groupby('Maternal Smoker')['Shuffled Birth Weight'].plot(kind='kde', title=title, ax=axes[0])
title = 'Birth Weights by Maternal Smoker'
original_and_shuffled.groupby('Maternal Smoker')['Birth Weight'].plot(kind='kde', title=title, ax=axes[1]);
n_repetitions = 500
differences = []
for _ in range(n_repetitions):
# Step 1: Shuffle the weights
shuffled_weights = (
smoking_and_birthweight['Birth Weight']
.sample(frac=1)
.reset_index(drop=True) # Be sure to reset the index! (Why?)
)
# Step 2: Put them in a DataFrame
shuffled = (
smoking_and_birthweight
.assign(**{'Shuffled Birth Weight': shuffled_weights})
)
# Step 3: Compute the test statistic
group_means = (
shuffled
.groupby('Maternal Smoker')
.mean()
.loc[:, 'Shuffled Birth Weight']
)
difference = group_means.diff().iloc[-1]
# Step 4: Store the result
differences.append(difference)
differences[:10]
[-0.03683593095357196, 0.6106464341758482, -0.5090330149153601, -2.6160336395630566, -1.90058351234822, 0.7644682115270456, -0.2478937184819614, -0.5483827719121876, -0.8667580785227926, 0.5069061657297027]
We already computed the observed statistic earlier, but we compute it again below to keep all of our calculations together.
observed_difference = (
smoking_and_birthweight
.groupby('Maternal Smoker')['Birth Weight']
.mean()
.diff()
.iloc[-1]
)
observed_difference
-9.266142572024918
title = 'Mean Differences in Birth Weights (Smoker - Non-Smoker)'
pd.Series(differences).plot(kind='hist', density=True, ec='w', bins=10, title=title)
plt.axvline(x=observed_difference, color='red', linewidth=3);
couples_fp = os.path.join('data', 'married_couples.csv')
couples = pd.read_csv(couples_fp)
couples.head()
| hh_id | gender | mar_status | rel_rating | age | education | hh_income | empl_status | hh_internet | |
|---|---|---|---|---|---|---|---|---|---|
| 0 | 0 | 1 | 1 | 1 | 51 | 12 | 14 | 1 | 1 |
| 1 | 0 | 2 | 1 | 1 | 53 | 9 | 14 | 1 | 1 |
| 2 | 1 | 1 | 1 | 1 | 57 | 11 | 15 | 1 | 1 |
| 3 | 1 | 2 | 1 | 1 | 57 | 9 | 15 | 1 | 1 |
| 4 | 2 | 1 | 1 | 1 | 60 | 12 | 14 | 1 | 1 |
We won't use all of the columns in the DataFrame.
couples = couples[['mar_status', 'empl_status', 'gender', 'age']]
couples.head()
| mar_status | empl_status | gender | age | |
|---|---|---|---|---|
| 0 | 1 | 1 | 1 | 51 |
| 1 | 1 | 1 | 2 | 53 |
| 2 | 1 | 1 | 1 | 57 |
| 3 | 1 | 1 | 2 | 57 |
| 4 | 1 | 1 | 1 | 60 |
The numbers in the DataFrame correspond to the mappings below.
mar_status: 1=married, 2=unmarried.empl_status: enumerated in the list below.gender: 1=male, 2=female.age: person's age in years.couples.head()
| mar_status | empl_status | gender | age | |
|---|---|---|---|---|
| 0 | 1 | 1 | 1 | 51 |
| 1 | 1 | 1 | 2 | 53 |
| 2 | 1 | 1 | 1 | 57 |
| 3 | 1 | 1 | 2 | 57 |
| 4 | 1 | 1 | 1 | 60 |
empl = [
'Working as paid employee',
'Working, self-employed',
'Not working - on a temporary layoff from a job',
'Not working - looking for work',
'Not working - retired',
'Not working - disabled',
'Not working - other'
]
couples = couples.replace({
'mar_status': {1:'married', 2:'unmarried'},
'gender': {1: 'M', 2: 'F'},
'empl_status': {(k + 1): empl[k] for k in range(len(empl))}
})
couples.head()
| mar_status | empl_status | gender | age | |
|---|---|---|---|---|
| 0 | married | Working as paid employee | M | 51 |
| 1 | married | Working as paid employee | F | 53 |
| 2 | married | Working as paid employee | M | 57 |
| 3 | married | Working as paid employee | F | 57 |
| 4 | married | Working as paid employee | M | 60 |
couples dataset¶# This cell shows the top 10 most common values in each column, along with their frequencies.
for col in couples:
print(col)
empr = couples[col].value_counts(normalize=True).to_frame().iloc[:10]
display(empr)
mar_status
| mar_status | |
|---|---|
| married | 0.717602 |
| unmarried | 0.282398 |
empl_status
| empl_status | |
|---|---|
| Working as paid employee | 0.605899 |
| Not working - other | 0.103965 |
| Working, self-employed | 0.098646 |
| Not working - looking for work | 0.067698 |
| Not working - disabled | 0.056576 |
| Not working - retired | 0.050774 |
| Not working - on a temporary layoff from a job | 0.016441 |
gender
| gender | |
|---|---|
| M | 0.5 |
| F | 0.5 |
age
| age | |
|---|---|
| 53 | 0.037234 |
| 55 | 0.036750 |
| 54 | 0.031431 |
| 40 | 0.030464 |
| 44 | 0.029981 |
| 30 | 0.028046 |
| 48 | 0.027563 |
| 49 | 0.027079 |
| 52 | 0.027079 |
| 43 | 0.026596 |
Ages are numeric, so the previous summary was not that helpful. Let's draw a histogram.
couples['age'].plot(kind='hist', density=True, ec='w', bins=np.arange(18, 66));
Let's look at the distribution of age separately for married couples and unmarried couples.
G = couples.groupby('mar_status')
ax = G.get_group('married')['age'].rename('Married').plot(kind='hist', density=True, alpha=0.75,
ec='w', bins=np.arange(18, 66, 2),
legend=True, title='Distribution of Ages (Married vs. Unmarried)')
G.get_group('unmarried')['age'].rename('Unmarried').plot(kind='hist', density=True, alpha=0.75,
ec='w', bins=np.arange(18, 66, 2),
ax=ax, legend=True);
What's the difference in the two distributions? Why do you think there is a difference?
To answer these questions, let's compute the distribution of employment status conditional on household type (married vs. unmarried).
couples.head()
| mar_status | empl_status | gender | age | |
|---|---|---|---|---|
| 0 | married | Working as paid employee | M | 51 |
| 1 | married | Working as paid employee | F | 53 |
| 2 | married | Working as paid employee | M | 57 |
| 3 | married | Working as paid employee | F | 57 |
| 4 | married | Working as paid employee | M | 60 |
# Note that this is a shortcut to picking a column for values and using aggfunc='count'
empl_cnts = couples.pivot_table(index='empl_status', columns='mar_status', aggfunc='size')
empl_cnts
| mar_status | married | unmarried |
|---|---|---|
| empl_status | ||
| Not working - disabled | 72 | 45 |
| Not working - looking for work | 71 | 69 |
| Not working - on a temporary layoff from a job | 21 | 13 |
| Not working - other | 182 | 33 |
| Not working - retired | 94 | 11 |
| Working as paid employee | 906 | 347 |
| Working, self-employed | 138 | 66 |
Since there are a different number of married and unmarried couples in the dataset, we can't compare the numbers above directly. We need to convert counts to proportions, separately for married and unmarried couples.
empl_cnts.sum()
mar_status married 1484 unmarried 584 dtype: int64
cond_distr = empl_cnts / empl_cnts.sum()
cond_distr
| mar_status | married | unmarried |
|---|---|---|
| empl_status | ||
| Not working - disabled | 0.048518 | 0.077055 |
| Not working - looking for work | 0.047844 | 0.118151 |
| Not working - on a temporary layoff from a job | 0.014151 | 0.022260 |
| Not working - other | 0.122642 | 0.056507 |
| Not working - retired | 0.063342 | 0.018836 |
| Working as paid employee | 0.610512 | 0.594178 |
| Working, self-employed | 0.092992 | 0.113014 |
Both of the columns above sum to 1.
cond_distr.plot(kind='barh', title='Distribution of Employment Status, Conditional on Household Type');
Null hypothesis: In the US, the distribution of employment status among those who are married is the same as among those who are unmarried and live with their partners. The difference between the two observed samples is due to chance.
Alternative hypothesis: In the US, the distributions of employment status of the two groups are different.
What is a good test statistic in this case?
Hint: What kind of distributions are we comparing?
cond_distr
| mar_status | married | unmarried |
|---|---|---|
| empl_status | ||
| Not working - disabled | 0.048518 | 0.077055 |
| Not working - looking for work | 0.047844 | 0.118151 |
| Not working - on a temporary layoff from a job | 0.014151 | 0.022260 |
| Not working - other | 0.122642 | 0.056507 |
| Not working - retired | 0.063342 | 0.018836 |
| Working as paid employee | 0.610512 | 0.594178 |
| Working, self-employed | 0.092992 | 0.113014 |
Let's first compute the observed TVD.
cond_distr.diff(axis=1).iloc[:, -1].abs().sum() / 2
0.1269754089281099
Since we'll need to calculate the TVD repeatedly, let's define a function that computes it.
couples.head()
| mar_status | empl_status | gender | age | |
|---|---|---|---|---|
| 0 | married | Working as paid employee | M | 51 |
| 1 | married | Working as paid employee | F | 53 |
| 2 | married | Working as paid employee | M | 57 |
| 3 | married | Working as paid employee | F | 57 |
| 4 | married | Working as paid employee | M | 60 |
def tvd_of_groups(df):
cnts = df.pivot_table(index='empl_status', columns='mar_status', aggfunc='size')
distr = cnts / cnts.sum() # Normalized
return distr.diff(axis=1).iloc[:, -1].abs().sum() / 2 # TVD
# Same result as above
observed_tvd = tvd_of_groups(couples)
observed_tvd
0.1269754089281099
couples.head()
| mar_status | empl_status | gender | age | |
|---|---|---|---|---|
| 0 | married | Working as paid employee | M | 51 |
| 1 | married | Working as paid employee | F | 53 |
| 2 | married | Working as paid employee | M | 57 |
| 3 | married | Working as paid employee | F | 57 |
| 4 | married | Working as paid employee | M | 60 |
Again, let's first figure out how to perform a single shuffle. Here, we'll shuffle marital statuses.
s = couples['mar_status'].sample(frac=1).reset_index(drop=True)
s
0 unmarried
1 married
2 married
3 married
4 married
...
2063 married
2064 married
2065 married
2066 married
2067 unmarried
Name: mar_status, Length: 2068, dtype: object
shuffled = couples.loc[:, ['empl_status']].assign(mar_status=s)
shuffled
| empl_status | mar_status | |
|---|---|---|
| 0 | Working as paid employee | unmarried |
| 1 | Working as paid employee | married |
| 2 | Working as paid employee | married |
| 3 | Working as paid employee | married |
| 4 | Working as paid employee | married |
| ... | ... | ... |
| 2063 | Working as paid employee | married |
| 2064 | Working as paid employee | married |
| 2065 | Working as paid employee | married |
| 2066 | Working as paid employee | married |
| 2067 | Working as paid employee | unmarried |
2068 rows × 2 columns
tvd_of_groups(shuffled)
0.06320385481667465
Let's do this repeatedly.
N = 1000
tvds = []
for _ in range(N):
s = couples['mar_status'].sample(frac=1).reset_index(drop=True)
shuffled = couples.loc[:, ['empl_status']].assign(mar_status=s)
tvds.append(tvd_of_groups(shuffled))
tvds = pd.Series(tvds)
Notice that by defining a function that computes our test statistic, our simulation code is much cleaner.
pval = (tvds >= observed_tvd).sum() / N
tvds.plot(kind='hist', density=True, ec='w', bins=20, title=f'P-value: {pval}', label='Simulated TVDs')
plt.axvline(x=observed_tvd, color='red', linewidth=3, label='P-value')
perc = np.percentile(tvds, 95) # 5% significance level
plt.axvline(x=perc, color='y', linewidth=3, label='P-value cutoff')
plt.legend();
In the definition of the TVD, we divide the sum of the absolute differences in proportions between the two distributions by 2.
def tvd(a, b):
return np.sum(np.abs(a - b)) / 2
Question: If we divided by 200 instead of 2, would we still reject the null hypothesis?
'Not working', but either 'looking for work' or 'disabled'?'Not working – looking for work' + 'Not working – disabled' individuals.couples.head()
| mar_status | empl_status | gender | age | |
|---|---|---|---|---|
| 0 | married | Working as paid employee | M | 51 |
| 1 | married | Working as paid employee | F | 53 |
| 2 | married | Working as paid employee | M | 57 |
| 3 | married | Working as paid employee | F | 57 |
| 4 | married | Working as paid employee | M | 60 |
couples['empl_status'].value_counts()
Working as paid employee 1253 Not working - other 215 Working, self-employed 204 Not working - looking for work 140 Not working - disabled 117 Not working - retired 105 Not working - on a temporary layoff from a job 34 Name: empl_status, dtype: int64
The Series isin method will be helpful here.
not_work_no_choice = couples['empl_status'].isin(['Not working - looking for work', 'Not working - disabled'])
not_work_no_choice
0 False
1 False
2 False
3 False
4 False
...
2063 False
2064 False
2065 False
2066 False
2067 False
Name: empl_status, Length: 2068, dtype: bool
couples['not_work_no_choice'] = not_work_no_choice.replace({True: 1, False: 0})
couples.head(12)
| mar_status | empl_status | gender | age | not_work_no_choice | |
|---|---|---|---|---|---|
| 0 | married | Working as paid employee | M | 51 | 0 |
| 1 | married | Working as paid employee | F | 53 | 0 |
| 2 | married | Working as paid employee | M | 57 | 0 |
| 3 | married | Working as paid employee | F | 57 | 0 |
| 4 | married | Working as paid employee | M | 60 | 0 |
| 5 | married | Working as paid employee | F | 57 | 0 |
| 6 | married | Working, self-employed | M | 62 | 0 |
| 7 | married | Working as paid employee | F | 59 | 0 |
| 8 | married | Not working - other | M | 53 | 0 |
| 9 | married | Not working - retired | F | 61 | 0 |
| 10 | married | Not working - on a temporary layoff from a job | M | 34 | 0 |
| 11 | married | Not working - looking for work | F | 32 | 1 |
Let's group by mar_status once again.
couples.groupby('mar_status')['not_work_no_choice'].mean()
mar_status married 0.096361 unmarried 0.195205 Name: not_work_no_choice, dtype: float64
Notice this is not a cateogrical distribution, so we don't need to use the TVD. Instead, we can just compute the difference in group means.
obs_mean = couples.groupby('mar_status')['not_work_no_choice'].mean().diff().iloc[-1]
obs_mean
0.0988442934682273
N = 1000
means = []
for _ in range(N):
s = couples['mar_status'].sample(frac=1).reset_index(drop=True)
shuffled = couples.loc[:, ['not_work_no_choice']].assign(mar_status=s)
m = shuffled.groupby('mar_status')['not_work_no_choice'].mean().diff().iloc[-1]
means.append(m)
means = pd.Series(means)
pval = (means >= obs_mean).sum() / N
means.plot(kind='hist', density=True, ec='w', bins=20, title=f'P-value: {pval}', label='Simulated Differences in Means')
plt.axvline(x=obs_mean, color='red', linewidth=3, label='P-value')
perc = np.percentile(means, 95) # 5% significance level
plt.axvline(x=perc, color='y', linewidth=3, label='P-value cutoff')
plt.legend();
Again, we reject the null hypothesis that married/unmarried households are similarly composed of those not working (not by choice) and otherwise.