Example: One-hot encoding 'STATE'¶
- For each unique value of
'STATE'in our dataset, we must create a column for just that'STATE'.
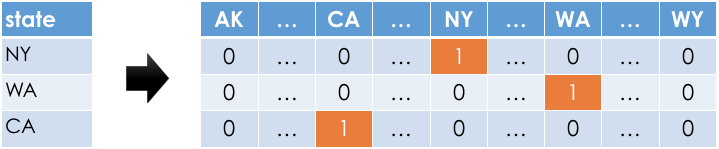
- Observations:
- In any given row, only one of the one-hot-encoded columns will contain a 1; the rest will contain a 0.
- Most of the values in the one-hot-encoded columns are 0, i.e. these columns are sparse.