import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
import seaborn as sns
# Carryover setup from last lecture
from sklearn.linear_model import LinearRegression
tips = sns.load_dataset('tips')
plt.style.use('seaborn-white')
plt.rc('figure', dpi=100, figsize=(7, 5))
plt.rc('font', size=12)
import warnings
warnings.simplefilter('ignore')

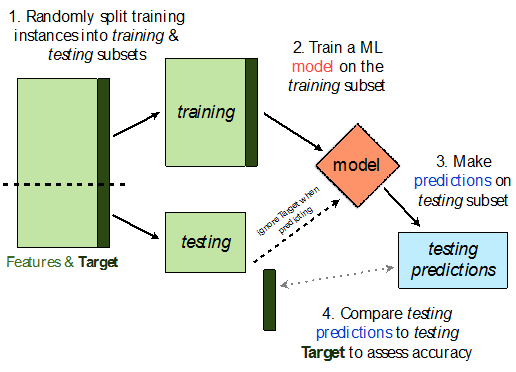
sklearn.model_selection.train_test_split implements a train-test split for us! 🙏🏼
If X is an array/DataFrame of features and y is an array/Series of responses,
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.25)
randomly splits the features and responses into training and test sets, such that the test set contains 0.25 of the full dataset.
from sklearn.model_selection import train_test_split
# Read the documentation!
train_test_split?
Let's perform a train/test split on our tips dataset.
X = tips.drop('tip', axis=1)
y = tips['tip']
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.2) # Don't have to choose 0.25
Before proceeding, let's check the sizes of X_train and X_test.
print('Rows in X_train:', X_train.shape[0])
display(X_train.head())
print('Rows in X_test:', X_test.shape[0])
display(X_test.head())
Rows in X_train: 195
| total_bill | sex | smoker | day | time | size | |
|---|---|---|---|---|---|---|
| 230 | 24.01 | Male | Yes | Sat | Dinner | 4 |
| 11 | 35.26 | Female | No | Sun | Dinner | 4 |
| 78 | 22.76 | Male | No | Thur | Lunch | 2 |
| 170 | 50.81 | Male | Yes | Sat | Dinner | 3 |
| 41 | 17.46 | Male | No | Sun | Dinner | 2 |
Rows in X_test: 49
| total_bill | sex | smoker | day | time | size | |
|---|---|---|---|---|---|---|
| 186 | 20.90 | Female | Yes | Sun | Dinner | 3 |
| 228 | 13.28 | Male | No | Sat | Dinner | 2 |
| 188 | 18.15 | Female | Yes | Sun | Dinner | 3 |
| 234 | 15.53 | Male | Yes | Sat | Dinner | 2 |
| 239 | 29.03 | Male | No | Sat | Dinner | 3 |
X_train.shape[0] / tips.shape[0]
0.7991803278688525
Steps:
tips.head()
| total_bill | tip | sex | smoker | day | time | size | |
|---|---|---|---|---|---|---|---|
| 0 | 16.99 | 1.01 | Female | No | Sun | Dinner | 2 |
| 1 | 10.34 | 1.66 | Male | No | Sun | Dinner | 3 |
| 2 | 21.01 | 3.50 | Male | No | Sun | Dinner | 3 |
| 3 | 23.68 | 3.31 | Male | No | Sun | Dinner | 2 |
| 4 | 24.59 | 3.61 | Female | No | Sun | Dinner | 4 |
X = tips[['total_bill', 'size']] # For this example, we'll use just the already-quantitative columns in tips
y = tips['tip']
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.2, random_state=1)
Here, we'll use a stand-alone LinearRegression model without a Pipeline, but this process would work the same if we were using a Pipeline.
lr = LinearRegression()
lr.fit(X_train, y_train)
LinearRegression()
Let's check our model's performance on the training set first.
from sklearn.metrics import mean_squared_error # built-in RMSE/MSE function
pred_train = lr.predict(X_train)
rmse_train = mean_squared_error(y_train, pred_train, squared=False)
rmse_train
0.9803205287924736
And the test set:
pred_test = lr.predict(X_test)
rmse_test = mean_squared_error(y_test, pred_test, squared=False)
rmse_test
1.138177129113125
Since rmse_train and rmse_test are similar, it doesn't seem like our model is overfitting to the training data. If rmse_test was much larger than rmse_train, it would be evidence that our model is unable to generalize well.
Recall, last class we looked at an example of polynomial regression.
When building these models:
A parameter defines the relationship between variables in a model.
For instance, suppose we fit a degree 3 polynomial to data, and end up with
$$H(x) = 1 - 2x + 13x^2 - 4x^3$$
1, -2, 13, and -4 are parameters.
sample_1 = pd.read_csv('data/sample-1.csv')
sample_1.head()
| x | y | |
|---|---|---|
| 0 | -2.000000 | -5.999036 |
| 1 | -1.949495 | -7.331676 |
| 2 | -1.898990 | -9.180925 |
| 3 | -1.848485 | -3.470179 |
| 4 | -1.797980 | -3.707370 |
plt.scatter(sample_1['x'], sample_1['y']);
First, we perform a train-test split.
X = sample_1[['x']]
y = sample_1['y']
X_train, X_test, y_train, y_test = train_test_split(X, y, random_state=100) # random_state is like np.random.seed
Now, we'll implement the logic from the previous slide.
from sklearn.pipeline import Pipeline
from sklearn.preprocessing import PolynomialFeatures
train_errs = []
test_errs = []
for d in range(1, 16):
pl = Pipeline([('poly', PolynomialFeatures(d)), ('lin-reg', LinearRegression())])
pl.fit(X_train, y_train)
train_errs.append(mean_squared_error(y_train, pl.predict(X_train), squared=False))
test_errs.append(mean_squared_error(y_test, pl.predict(X_test), squared=False))
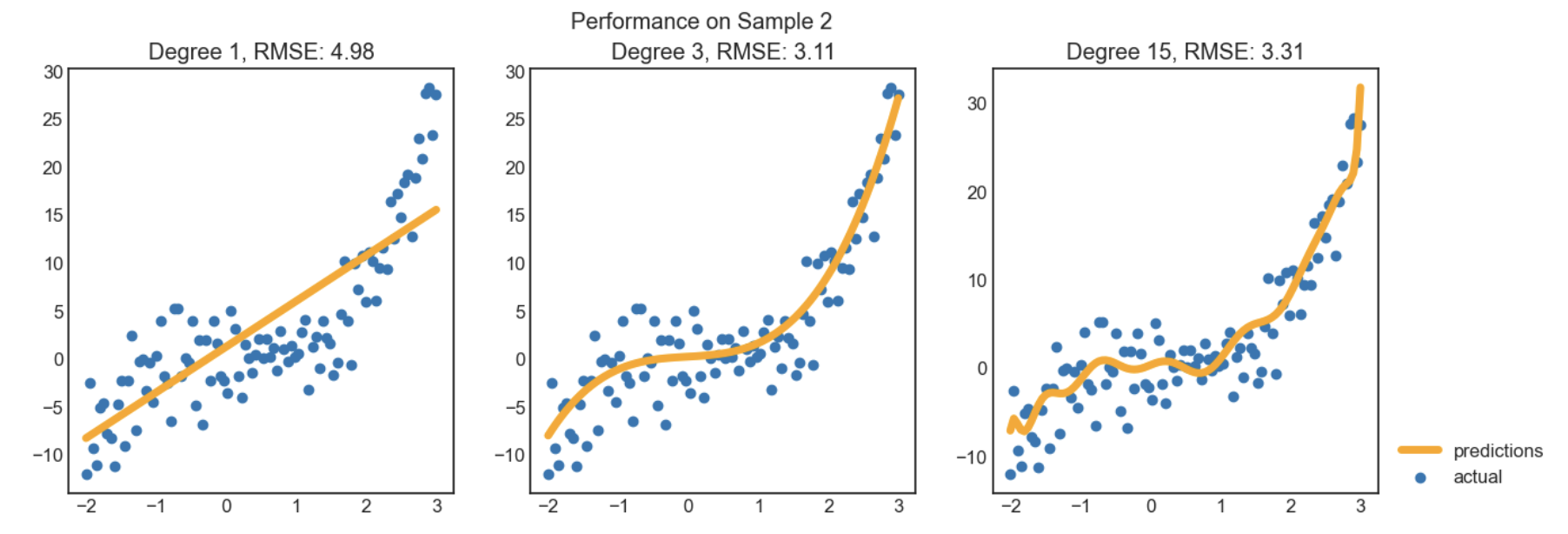
Let's look at both the training RMSEs and test RMSEs we computed.
plt.plot(range(1, 16), train_errs, label='training RMSE')
plt.plot(range(1, 16), test_errs, label='test RMSE')
plt.xlabel('Polynomial Degree')
plt.ylabel('RMSE')
plt.legend();
Observations:
Here, we'd choose a degree of 3, since that degree has the lowest test error.
We pick the hyperparameter(s) at the "valley" of test error.
Note that training error tends to underestimate test error, but it doesn't have to – i.e., it is possible for test error to be lower than training error (say, if the test set is "easier" to predict than the training set).
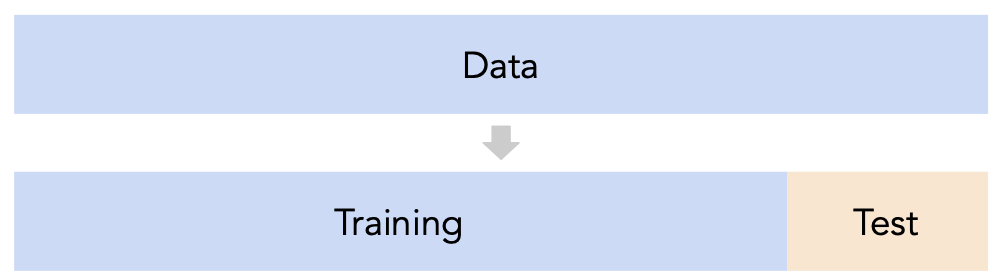
sklearn's train_test_split splits randomly, which usually works well.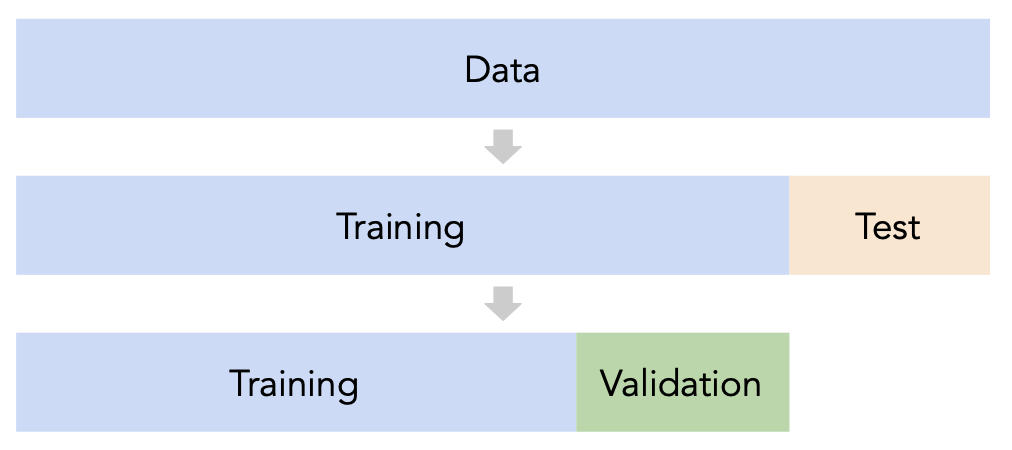
Issue: This strategy is too dependent on the validation set, which may be small and/or not a representative sample of the data.
Instead of relying on a single validation set, we can create $k$ validation sets, where $k$ is some positive integer (5 in the following example).
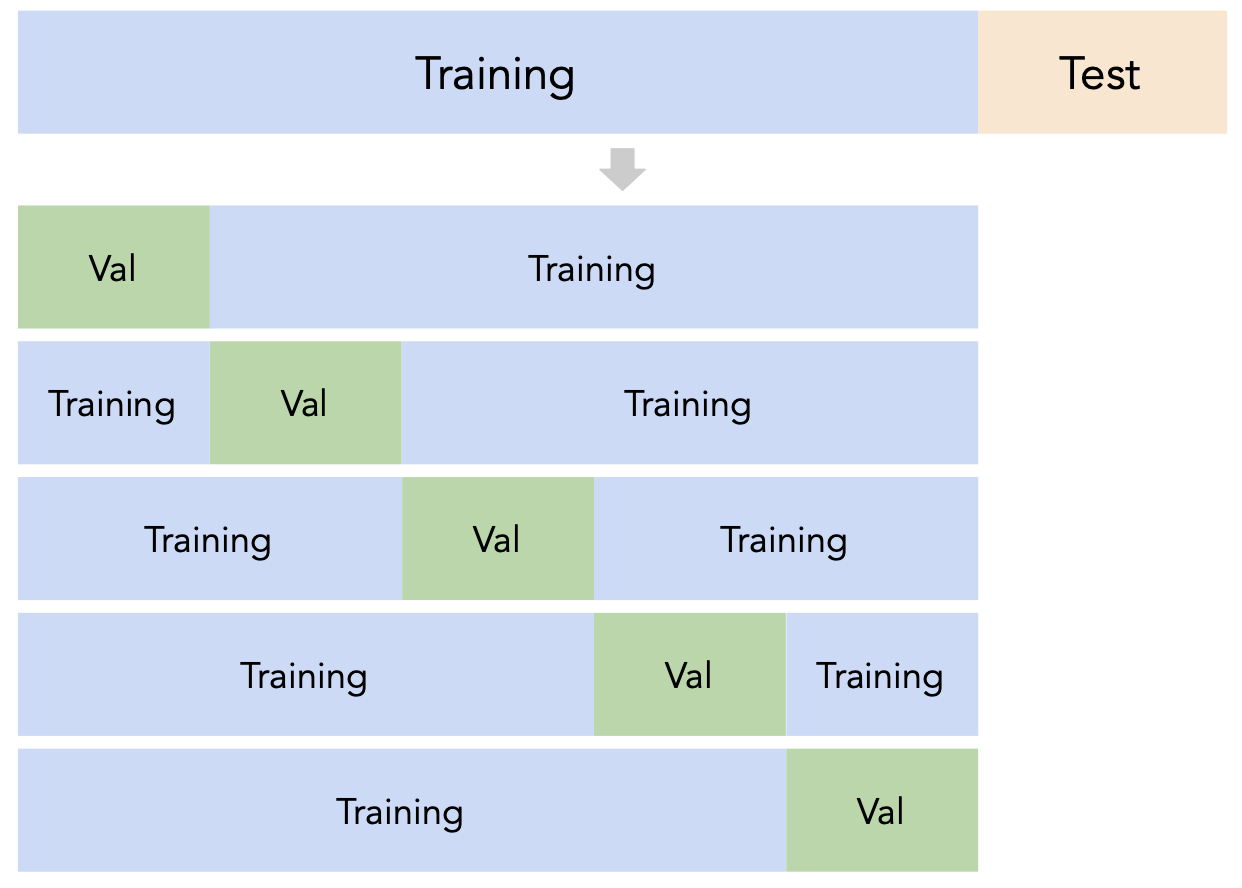
Since each data point is used for training $k-1$ times and validation once, the (averaged) validation performance should be a good metric of a model's ability to generalize to unseen data.
First, shuffle the dataset randomly and split it into $k$ disjoint groups. Then:
sklearn¶sklearn has a KFold class that splits data into training and validation folds.
from sklearn.model_selection import KFold
Let's use a simple dataset for illustration.
data = np.arange(10, 70, 10)
data
array([10, 20, 30, 40, 50, 60])
Let's instantiate a KFold object with $k=3$.
kfold = KFold(3, shuffle=True, random_state=1)
kfold
KFold(n_splits=3, random_state=1, shuffle=True)
Finally, let's use kfold to split data:
for train, val in kfold.split(data):
print(f'train: {data[train]}, validation: {data[val]}')
train: [10 40 50 60], validation: [20 30] train: [20 30 40 60], validation: [10 50] train: [10 20 30 50], validation: [40 60]
Note that each value in data is used for validation exactly once and for training exactly twice. Also note that because we set shuffle=True the groups are not simply [10, 20], [30, 40], and [50, 60].
sklearn¶KFold to perform $k$-fold cross validation in order to help us pick a degree for polynomial regression from the list [1, 2, ..., 15]. sklearn).sample_1 is our "training + validation data", i.e. that our test data is in some other dataset.sample_1 into train and test.plt.scatter(sample_1['x'], sample_1['y']);
kfold = KFold(5, shuffle=True, random_state=1)
errs_df = pd.DataFrame()
for d in range(1, 16):
errs = []
for train, val in kfold.split(sample_1):
# Separate the data into a training set and validation set
data_train, data_val = sample_1.iloc[train], sample_1.iloc[val]
# Fit the model on the training set
pl = Pipeline([('poly', PolynomialFeatures(d)), ('lin-reg', LinearRegression())])
pl.fit(data_train[['x']], data_train['y'])
# Compute the model's validation error
val_err = mean_squared_error(data_val['y'], pl.predict(data_val[['x']]), squared=False)
errs.append(val_err)
errs_df[f'Deg {d}'] = errs
errs_df.index = [f'Fold {i}' for i in range(1, 6)]
errs_df
| Deg 1 | Deg 2 | Deg 3 | Deg 4 | Deg 5 | Deg 6 | Deg 7 | Deg 8 | Deg 9 | Deg 10 | Deg 11 | Deg 12 | Deg 13 | Deg 14 | Deg 15 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Fold 1 | 4.199778 | 3.606615 | 2.831147 | 2.828540 | 2.841994 | 2.858593 | 3.024709 | 3.065179 | 3.064011 | 3.087646 | 3.069269 | 3.035684 | 3.158415 | 3.185333 | 3.267513 |
| Fold 2 | 6.332986 | 4.758864 | 3.359064 | 3.463889 | 4.039780 | 4.080516 | 4.156041 | 4.197769 | 4.251904 | 4.066354 | 3.876995 | 3.870712 | 3.795465 | 3.986883 | 3.487144 |
| Fold 3 | 4.513414 | 3.091568 | 2.824030 | 2.904488 | 3.078070 | 3.201268 | 3.207738 | 3.224326 | 3.249295 | 3.317992 | 3.282708 | 3.448616 | 3.507821 | 3.561160 | 3.492002 |
| Fold 4 | 4.554400 | 4.044258 | 3.048785 | 3.053034 | 3.148588 | 3.093417 | 3.104223 | 3.150751 | 3.322878 | 3.358298 | 3.373014 | 3.326871 | 3.529446 | 3.487267 | 4.238141 |
| Fold 5 | 4.285422 | 3.594867 | 2.636512 | 2.676925 | 2.709439 | 3.267328 | 3.271033 | 3.268576 | 3.398126 | 3.324115 | 3.484460 | 3.498631 | 3.499423 | 3.549985 | 3.391811 |
Note that for each choice of degree (our hyperparameter), we have five RMSEs, one for each "fold" of the data.
We should choose the degree with the lowest average validation RMSE.
errs_df.mean()
Deg 1 4.777200 Deg 2 3.819234 Deg 3 2.939908 Deg 4 2.985375 Deg 5 3.163574 Deg 6 3.300225 Deg 7 3.352749 Deg 8 3.381320 Deg 9 3.457243 Deg 10 3.430881 Deg 11 3.417289 Deg 12 3.436103 Deg 13 3.498114 Deg 14 3.554126 Deg 15 3.575322 dtype: float64
errs_df.mean().idxmin()
'Deg 3'
Note that if we only performed non-$k$-fold cross-validation, we might pick a different degree:
for fold in errs_df.index:
print(errs_df.loc[fold].idxmin())
Deg 4 Deg 3 Deg 3 Deg 3 Deg 3
sklearn¶The cross_val_score function in sklearn implements a few of the previous steps in one.
cross_val_score(estimator, data, target, cv)
Specifically, it takes in:
Pipeline or estimator that has not already been fitcv argument)scoring metricand performs $k$-fold cross-validation, returning the values of the scoring metric on each fold.
from sklearn.model_selection import cross_val_score
errs_df_auto = pd.DataFrame()
for d in range(1, 16):
pl = Pipeline([('poly', PolynomialFeatures(d)), ('lin-reg', LinearRegression())])
# The `scoring` argument is used to specify that we want to compute the RMSE; the default is R^2
# It is called "neg" RMSE because by default sklearn likes to "maximize" scores
errs = cross_val_score(pl, sample_1[['x']], sample_1['y'], cv=5, scoring='neg_root_mean_squared_error')
errs_df_auto[f'Deg {d}'] = -errs # Negate to turn positive (sklearn computed negative RMSE)
errs_df_auto.index = [f'Fold {i}' for i in range(1, 6)]
errs_df_auto
| Deg 1 | Deg 2 | Deg 3 | Deg 4 | Deg 5 | Deg 6 | Deg 7 | Deg 8 | Deg 9 | Deg 10 | Deg 11 | Deg 12 | Deg 13 | Deg 14 | Deg 15 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Fold 1 | 4.789975 | 12.814865 | 5.040947 | 4.927487 | 8.853939 | 13.150006 | 18.023855 | 149.574851 | 897.759319 | 718.403458 | 841.062760 | 3964.733244 | 55940.500810 | 55106.074263 | 202369.608764 |
| Fold 2 | 3.971600 | 5.359234 | 3.186076 | 3.218728 | 3.194274 | 3.007987 | 3.077643 | 3.127204 | 3.098704 | 3.559998 | 4.260110 | 5.223040 | 4.996440 | 8.254853 | 6.843680 |
| Fold 3 | 4.770176 | 2.557932 | 2.083676 | 2.110708 | 2.100757 | 2.013973 | 2.018524 | 1.996626 | 2.186183 | 3.946495 | 3.889999 | 3.962834 | 4.021369 | 4.912291 | 4.772074 |
| Fold 4 | 6.134098 | 4.656187 | 2.925225 | 2.932392 | 3.030811 | 3.040322 | 3.144730 | 3.160528 | 3.310559 | 3.070833 | 2.931583 | 4.200841 | 3.894982 | 5.317150 | 3.238713 |
| Fold 5 | 11.699003 | 11.916631 | 3.235882 | 4.372698 | 33.584482 | 51.745272 | 56.510688 | 92.223491 | 335.804723 | 1168.510898 | 1128.161798 | 8074.430848 | 8910.388508 | 9651.165907 | 139023.154682 |
errs_df_auto.mean().idxmin()
'Deg 3'
That was considerably easier! Next class, we'll look at how to streamline this procedure even more (no loop necessary).
Note: You may notice that the RMSEs in the above table, particularly in Folds 1 and 5, are much higher than they were in the manual method. Can you think of reasons why, and how we might fix this? (Hint: Go back to the "manual" method and switch shuffle to False. What do you notice?)
Split the data into two sets: training and test.
Use only the training data when designing, training, and tuning the model.
Commit to your final model and train it using the entire training set.
Test the data using the test data. If the performance (e.g. RMSE) is not acceptable, return to step 2.
Finally, train on all available data and ship the model to production! 🛳
🚨 This is the process you should always use! 🚨
X.iloc[0]) used for training a model?
Decision trees can be used for both regression and classification. We will start by discussing their use in classification.
diabetes = pd.read_csv('data/diabetes.csv')
diabetes.head()
| Pregnancies | Glucose | BloodPressure | SkinThickness | Insulin | BMI | DiabetesPedigreeFunction | Age | Outcome | |
|---|---|---|---|---|---|---|---|---|---|
| 0 | 6 | 148 | 72 | 35 | 0 | 33.6 | 0.627 | 50 | 1 |
| 1 | 1 | 85 | 66 | 29 | 0 | 26.6 | 0.351 | 31 | 0 |
| 2 | 8 | 183 | 64 | 0 | 0 | 23.3 | 0.672 | 32 | 1 |
| 3 | 1 | 89 | 66 | 23 | 94 | 28.1 | 0.167 | 21 | 0 |
| 4 | 0 | 137 | 40 | 35 | 168 | 43.1 | 2.288 | 33 | 1 |
diabetes['Outcome'].value_counts()
0 500 1 268 Name: Outcome, dtype: int64
For illustration, we'll use 'Glucose' and 'BMI' to predict whether or not a patient has diabetes (the response variable is in the 'Outcome' column).
Let's build a decision tree and interpret the results. But first, a train-test split:
X_train, X_test, y_train, y_test = train_test_split(diabetes[['Glucose', 'BMI']],
diabetes['Outcome'],
random_state=1)
The relevant class is DecisionTreeClassifier, from sklearn.tree.
from sklearn.tree import DecisionTreeClassifier
Note that we fit it the same way we fit earlier estimators.
_You may wonder what max_depth=2 does – more on this soon!_
dt = DecisionTreeClassifier(max_depth=2)
dt.fit(X_train, y_train)
DecisionTreeClassifier(max_depth=2)
Our fit decision tree is like a "flowchart", made up of a series of questions.
Class 0 (orange) is "no diabetes"; Class 1 (blue) is "diabetes".
from sklearn.tree import plot_tree
plt.figure(figsize=(10, 5))
plot_tree(dt, feature_names=X_train.columns, class_names=['no', 'yes'],
filled=True, rounded=True, fontsize=15, impurity=False);
value attribute, which describes the number of training individuals of each class that fell in that node.# Note that the left node at depth 2 has a `value` of [304, 78]
y_train.loc[X_train[X_train['Glucose'] <= 129.5].index].value_counts()
0 304 1 78 Name: Outcome, dtype: int64
The most common evaluation metric in classification is accuracy:
$$\text{accuracy} = \frac{\text{# data points classified correctly}}{\text{# data points}}$$(dt.predict(X_train) == y_train).mean()
0.765625
The score method of a classifier computes accuracy by default.
# Training accuracy – same number as above
dt.score(X_train, y_train)
0.765625
# Testing accuracy
dt.score(X_test, y_test)
0.7760416666666666
sklearn decide what questions to ask?The answers will come next class!