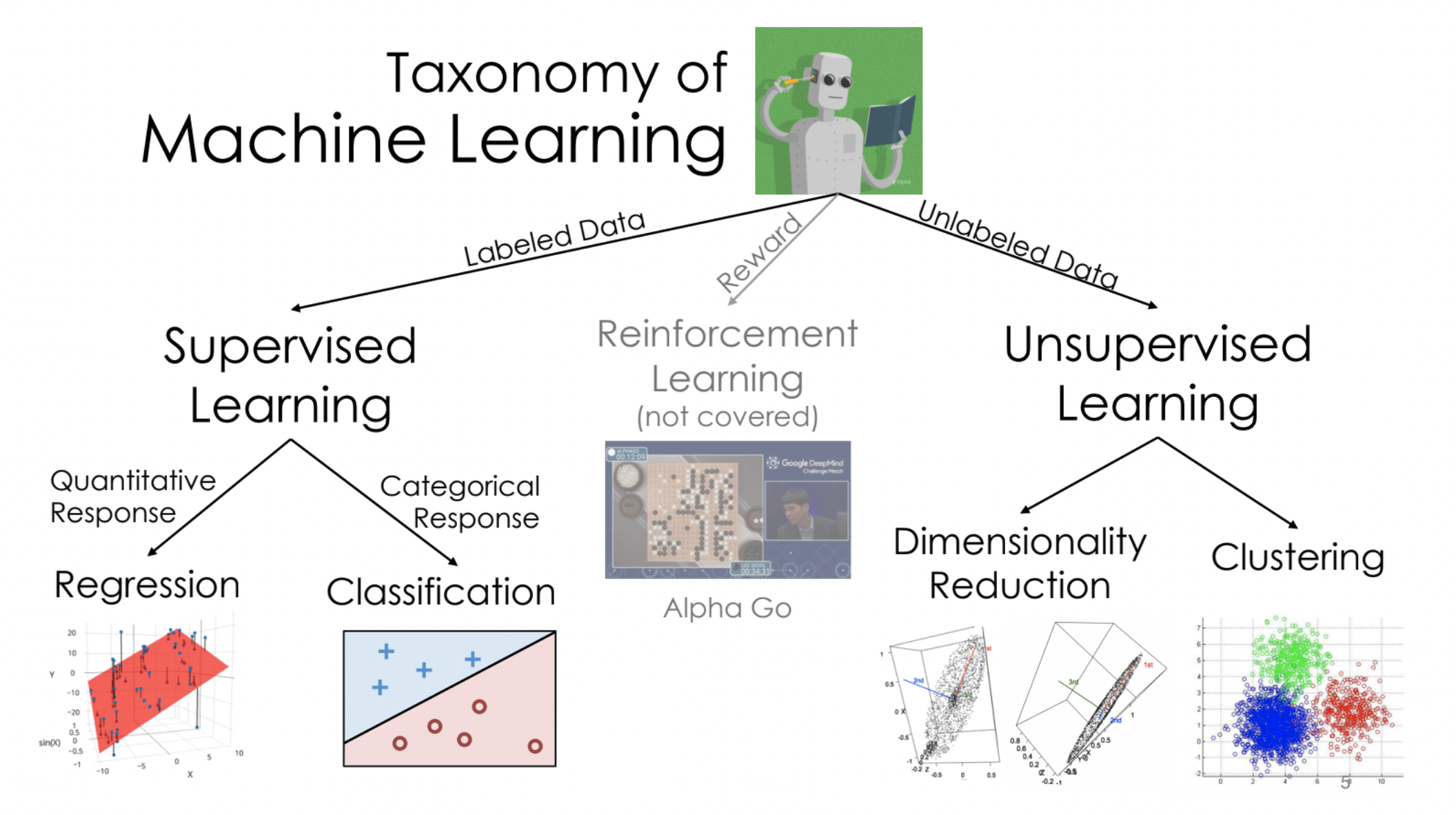
Decision trees can be used for both regression and classification. We will start by discussing their use in classification.
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
import seaborn as sns
from sklearn.model_selection import train_test_split
plt.style.use('seaborn-white')
plt.rc('figure', dpi=100, figsize=(7, 5))
plt.rc('font', size=12)
import warnings
warnings.simplefilter('ignore')
Split the data into two sets: training and test.
Use only the training data when designing, training, and tuning the model.
Commit to your final model and train it using the entire training set.
Test the data using the test data. If the performance (e.g. RMSE) is not acceptable, return to step 2.
Finally, train on all available data and ship the model to production! 🛳
🚨 This is the process you should always use! 🚨
We won't answer this question in class, but it's a good exam-prep question!
X.iloc[0]) used for training a model?
Decision trees can be used for both regression and classification. We will start by discussing their use in classification.
diabetes = pd.read_csv('data/diabetes.csv')
diabetes.head()
| Pregnancies | Glucose | BloodPressure | SkinThickness | Insulin | BMI | DiabetesPedigreeFunction | Age | Outcome | |
|---|---|---|---|---|---|---|---|---|---|
| 0 | 6 | 148 | 72 | 35 | 0 | 33.6 | 0.627 | 50 | 1 |
| 1 | 1 | 85 | 66 | 29 | 0 | 26.6 | 0.351 | 31 | 0 |
| 2 | 8 | 183 | 64 | 0 | 0 | 23.3 | 0.672 | 32 | 1 |
| 3 | 1 | 89 | 66 | 23 | 94 | 28.1 | 0.167 | 21 | 0 |
| 4 | 0 | 137 | 40 | 35 | 168 | 43.1 | 2.288 | 33 | 1 |
diabetes['Outcome'].value_counts()
0 500 1 268 Name: Outcome, dtype: int64
For illustration, we'll use 'Glucose' and 'BMI' to predict whether or not a patient has diabetes (the response variable is in the 'Outcome' column).
Let's build a decision tree and interpret the results. But first, a train-test split:
X_train, X_test, y_train, y_test = train_test_split(diabetes[['Glucose', 'BMI']],
diabetes['Outcome'],
random_state=1)
The relevant class is DecisionTreeClassifier, from sklearn.tree.
from sklearn.tree import DecisionTreeClassifier
Note that we fit it the same way we fit earlier estimators.
_You may wonder what max_depth=2 does – more on this soon!_
dt = DecisionTreeClassifier(max_depth=2)
dt.fit(X_train, y_train)
DecisionTreeClassifier(max_depth=2)
Our fit decision tree is like a "flowchart", made up of a series of questions.
Class 0 (orange) is "no diabetes"; Class 1 (blue) is "diabetes".
from sklearn.tree import plot_tree
plt.figure(figsize=(10, 5))
plot_tree(dt, feature_names=X_train.columns, class_names=['no', 'yes'],
filled=True, rounded=True, fontsize=15, impurity=False);
value attribute, which describes the number of training individuals of each class that fell in that node.# Note that the left node at depth 2 has a `value` of [304, 78]
y_train.loc[X_train[X_train['Glucose'] <= 129.5].index].value_counts()
0 304 1 78 Name: Outcome, dtype: int64
The most common evaluation metric in classification is accuracy:
$$\text{accuracy} = \frac{\text{# data points classified correctly}}{\text{# data points}}$$(dt.predict(X_train) == y_train).mean()
0.765625
The score method of a classifier computes accuracy by default.
# Training accuracy – same number as above
dt.score(X_train, y_train)
0.765625
# Testing accuracy
dt.score(X_test, y_test)
0.7760416666666666
sklearn decide what questions to ask?When we ask a question, we are effectively splitting a node into two children – the "yes" child and the "no" child.
Suppose the distribution within a node looks like this (colors represent classes):
Question A splits the node like this:
Question B splits the node like this:
Which question is "better"?
Question B, because there is "less uncertainty" in the resulting nodes after splitting by Question B than there is after splitting by Question A. There are two common techniques for quantifying "uncertainty":
sklearn).Not the focus of our course, but read more!
Decision trees are trained by recursively picking the best split until:
By default, there is no "maximum depth" for a decision tree. As such, without restriction, decision trees tend to be very deep.
dt_no_max = DecisionTreeClassifier()
dt_no_max.fit(X_train, y_train)
DecisionTreeClassifier()
A decision tree fit on our training data has a depth of around 20! (It is so deep that tree.plot_tree errors when trying to plot it.)
dt_no_max.tree_.max_depth
19
At first, this tree seems "better" than our tree of depth 2, since its training accuracy is much much higher:
dt_no_max.score(X_train, y_train)
0.9913194444444444
# Depth 2 tree
dt.score(X_train, y_train)
0.765625
But recall, we truly care about test set performance, and this decision tree has worse accuracy on the test set than our depth 2 tree.
dt_no_max.score(X_test, y_test)
0.7135416666666666
# Depth 2 tree
dt.score(X_test, y_test)
0.7760416666666666
Since sklearn.tree's plot_tree can't visualize extremely large decision trees, let's create and visualize some smaller decision trees.
trees = {}
for d in [2, 4, 8]:
trees[d] = DecisionTreeClassifier(max_depth=d, random_state=1)
trees[d].fit(X_train, y_train)
plt.figure(figsize=(15, 5), dpi=100)
plot_tree(trees[d], feature_names=X_train.columns, class_names=['no', 'yes'],
filled=True, rounded=True, impurity=False)
plt.show()
As tree depth increases, complexity increases.
Question: What is the right maximum depth to choose?
max_depth is a hyperparameter for DecisionTreeClassifier.min_samples_split: The minimum number of samples required to split an internal node.criterion: The function to measure the quality of a split ('gini' or 'entropy').GridSearchCV takes in:
fit instance of an estimator, andand performs $k$-fold cross-validation to find the combination of hyperparameters with the best average validation performance.
from sklearn.model_selection import GridSearchCV
The following dictionary contains the values we're considering for each hyperparameter. (We're using GridSearchCV with 3 hyperparameters, but we could it with even just a single hyperparameter.)
hyperparameters = {
'max_depth': [2, 3, 4, 5, 7, 10, 13, 15, 18, None],
'min_samples_split': [2, 3, 5, 7, 10, 15, 20],
'criterion': ['gini', 'entropy']
}
Note that there are 140 combinations of hyperparameters we need to try. We need to find the best combination of hyperparameters, not the best value for each hyperparameter individually.
len(hyperparameters['max_depth']) * len(hyperparameters['min_samples_split']) * len(hyperparameters['criterion'])
140
GridSearchCV needs to be instantiated and fit.
searcher = GridSearchCV(DecisionTreeClassifier(), hyperparameters, cv=5)
searcher.fit(X_train, y_train)
GridSearchCV(cv=5, estimator=DecisionTreeClassifier(),
param_grid={'criterion': ['gini', 'entropy'],
'max_depth': [2, 3, 4, 5, 7, 10, 13, 15, 18, None],
'min_samples_split': [2, 3, 5, 7, 10, 15, 20]})
After being fit, the best_params_ attribute provides us with the best combination of hyperparameters to use.
searcher.best_params_
{'criterion': 'gini', 'max_depth': 4, 'min_samples_split': 7}
All of the intermediate results – validation accuracies for each fold, mean validation accuaries, etc. – are stored in the cv_results_ attribute:
searcher.cv_results_['mean_test_score'] # array of length 140
array([0.7292054 , 0.7292054 , 0.7292054 , 0.7292054 , 0.7292054 ,
0.7292054 , 0.7292054 , 0.74136432, 0.74136432, 0.74136432,
0.74136432, 0.74136432, 0.74136432, 0.74136432, 0.75005997,
0.75005997, 0.75005997, 0.7517991 , 0.74832084, 0.7517991 ,
0.75005997, 0.73613193, 0.73787106, 0.73613193, 0.73961019,
0.73961019, 0.74134933, 0.73787106, 0.71364318, 0.71364318,
0.71881559, 0.724003 , 0.73097451, 0.73967016, 0.73617691,
0.70838081, 0.71182909, 0.70835082, 0.71875562, 0.72052474,
0.72748126, 0.7292054 , 0.68928036, 0.69101949, 0.69794603,
0.70833583, 0.70491754, 0.71362819, 0.71709145, 0.69275862,
0.70314843, 0.70833583, 0.7170015 , 0.72395802, 0.7188006 ,
0.71704648, 0.698006 , 0.69448276, 0.6961919 , 0.70482759,
0.70656672, 0.7188006 , 0.72226387, 0.69275862, 0.68584708,
0.70143928, 0.70661169, 0.70662669, 0.7188006 , 0.72226387,
0.72398801, 0.72398801, 0.72398801, 0.72398801, 0.72398801,
0.72398801, 0.72398801, 0.74658171, 0.74658171, 0.74658171,
0.74658171, 0.74658171, 0.74658171, 0.74658171, 0.7517991 ,
0.7517991 , 0.7517991 , 0.75005997, 0.75005997, 0.75005997,
0.75005997, 0.75004498, 0.75004498, 0.75178411, 0.75178411,
0.75178411, 0.75178411, 0.75178411, 0.72923538, 0.72922039,
0.7292054 , 0.73962519, 0.74482759, 0.75005997, 0.74656672,
0.71881559, 0.70665667, 0.72056972, 0.73095952, 0.73965517,
0.7414093 , 0.7413943 , 0.69802099, 0.69275862, 0.69626687,
0.71535232, 0.72227886, 0.73097451, 0.73443778, 0.69098951,
0.68406297, 0.69446777, 0.70835082, 0.71533733, 0.72923538,
0.73269865, 0.67890555, 0.68233883, 0.69445277, 0.69968516,
0.71011994, 0.73097451, 0.73269865, 0.67893553, 0.6858021 ,
0.68929535, 0.70316342, 0.70838081, 0.72752624, 0.73269865])
# Rows correspond to folds, columns correspond to hyperparameter combinations
pd.DataFrame(np.vstack([searcher.cv_results_[f'split{i}_test_score'] for i in range(5)]))
| 0 | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | ... | 130 | 131 | 132 | 133 | 134 | 135 | 136 | 137 | 138 | 139 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 0.706897 | 0.706897 | 0.706897 | 0.706897 | 0.706897 | 0.706897 | 0.706897 | 0.715517 | 0.715517 | 0.715517 | ... | 0.681034 | 0.689655 | 0.698276 | 0.612069 | 0.663793 | 0.655172 | 0.681034 | 0.681034 | 0.672414 | 0.698276 |
| 1 | 0.773913 | 0.773913 | 0.773913 | 0.773913 | 0.773913 | 0.773913 | 0.773913 | 0.756522 | 0.756522 | 0.756522 | ... | 0.747826 | 0.800000 | 0.817391 | 0.695652 | 0.678261 | 0.669565 | 0.713043 | 0.765217 | 0.800000 | 0.817391 |
| 2 | 0.739130 | 0.739130 | 0.739130 | 0.739130 | 0.739130 | 0.739130 | 0.739130 | 0.756522 | 0.756522 | 0.756522 | ... | 0.678261 | 0.704348 | 0.678261 | 0.669565 | 0.669565 | 0.678261 | 0.686957 | 0.686957 | 0.704348 | 0.678261 |
| 3 | 0.704348 | 0.704348 | 0.704348 | 0.704348 | 0.704348 | 0.704348 | 0.704348 | 0.756522 | 0.756522 | 0.756522 | ... | 0.747826 | 0.765217 | 0.765217 | 0.730435 | 0.721739 | 0.739130 | 0.739130 | 0.721739 | 0.765217 | 0.765217 |
| 4 | 0.721739 | 0.721739 | 0.721739 | 0.721739 | 0.721739 | 0.721739 | 0.721739 | 0.721739 | 0.721739 | 0.721739 | ... | 0.695652 | 0.695652 | 0.704348 | 0.686957 | 0.695652 | 0.704348 | 0.695652 | 0.686957 | 0.695652 | 0.704348 |
5 rows × 140 columns
Note that the above DataFrame tells us that 5 * 140 = 700 models were trained in total!
Question: How is the following line of code making predictions?
searcher.predict(X_train)[:5]
array([0, 0, 1, 0, 0])
Which model's testing accuracy is shown below?
searcher.score(X_test, y_test)
0.765625
GridSearchCV.GridSearchCV will try all unique combinations of hyperparameters and return the combination with the best average validation performance.GridSearchCV is not the only solution – see RandomizedSearchCV if you're curious.Suppose we fit a simple linear regression model that uses height in inches to predict weight in pounds.
$$\text{predicted weight} = w_0 + w_1 \cdot \text{height (inches)}$$people = pd.read_csv('data/SOCR-HeightWeight.csv').drop('Index', axis=1)
people.head()
| Height(Inches) | Weight(Pounds) | |
|---|---|---|
| 0 | 65.78331 | 112.9925 |
| 1 | 71.51521 | 136.4873 |
| 2 | 69.39874 | 153.0269 |
| 3 | 68.21660 | 142.3354 |
| 4 | 67.78781 | 144.2971 |
from sklearn.linear_model import LinearRegression
from sklearn.metrics import mean_squared_error
X_train_1, X_test_1, y_train_1, y_test_1 = train_test_split(people[['Height(Inches)']],
people['Weight(Pounds)'],
random_state=1)
lr_one_feat = LinearRegression()
lr_one_feat.fit(X_train_1, y_train_1)
LinearRegression()
$w_0^*$ and $w_1^*$ are shown below, along with the model's testing RMSE.
lr_one_feat.intercept_, lr_one_feat.coef_
(-81.18621383194537, array([3.06263359]))
rmse_one_feat = mean_squared_error(y_test_1,
lr_one_feat.predict(X_test_1),
squared=False)
rmse_one_feat
10.044723811394736
Now, suppose we fit another regression model, that uses height in inches AND height in centimeters to predict weight.
$$\text{predicted weight} = w_0 + w_1 \cdot \text{height (inches)} + w_2 \cdot \text{height (cm)}$$people['Height(cm)'] = people['Height(Inches)'] * 2.54 # 1 inch = 2.54 cm
X_train_2, X_test_2, y_train_2, y_test_2 = train_test_split(people[['Height(Inches)', 'Height(cm)']],
people['Weight(Pounds)'],
random_state=1)
lr_two_feat = LinearRegression()
lr_two_feat.fit(X_train_2, y_train_2)
LinearRegression()
What are $w_0^*$, $w_1^*$, $w_2^*$, and the model's testing RMSE?
lr_two_feat.intercept_, lr_two_feat.coef_
(-81.16916912853335, array([ 9.70933657e+11, -3.82257345e+11]))
rmse_two_feat = mean_squared_error(y_test_2,
lr_two_feat.predict(X_test_2),
squared=False)
rmse_two_feat
10.044733718499367
Observation: The intercept is the same as before (roughly -81.17), as is the testing RMSE. However, the coefficients on 'Height(Inches)' and 'Height(cm)' are massive in size!
What's going on?
Let's use simpler numbers for illustration. Suppose in the first model, $w_0^* = -80$ and $w_1^* = 3$.
In the second model, we have:
$$\begin{align*}\text{predicted weight} &= w_0^* + w_1^* \cdot \text{height (inches)} + w_2^* \cdot \text{height (cm)} \\ &= w_0^* + w_1^* \cdot \text{height (inches)} + w_2^* \cdot \big( 2.54^* \cdot \text{height (inches)} \big) \\ &= w_0^* + \left(w_1^* + 2.54 \cdot w_2^* \right) \cdot \text{height (inches)} \end{align*}$$In the first model, we already found the "best" intercept and slope in a linear model that uses height in inches to predict weight ($-80$ and $3$).
So, as long as $w_1^* + 2.54 \cdot w_2^* = 3$ in the second model, the second model's training predictions will be the same as the first, and hence they will also minimize RMSE.
Issue: There are an infinite number of $w_1^*$ and $w_2^*$ that satisfy $w_1^* + 2.54 \cdot w_2^* = 3$!
lr.coef_ could return either set of coefficients, or any other of the infinitely many options. (-80 - 10 * people.iloc[:, 0] + (13 / 2.54) * people.iloc[:, 2]).head()
0 117.34993 1 134.54563 2 128.19622 3 124.64980 4 123.36343 dtype: float64
(-80 + 10 * people.iloc[:, 0] - (7 / 2.54) * people.iloc[:, 2]).head()
0 117.34993 1 134.54563 2 128.19622 3 124.64980 4 123.36343 dtype: float64
When we one-hot encode categorical features, we create several redundant columns.
tips = sns.load_dataset('tips')
tips_features = tips.drop('tip', axis=1)
tips_features.head()
| total_bill | sex | smoker | day | time | size | |
|---|---|---|---|---|---|---|
| 0 | 16.99 | Female | No | Sun | Dinner | 2 |
| 1 | 10.34 | Male | No | Sun | Dinner | 3 |
| 2 | 21.01 | Male | No | Sun | Dinner | 3 |
| 3 | 23.68 | Male | No | Sun | Dinner | 2 |
| 4 | 24.59 | Female | No | Sun | Dinner | 4 |
Aside: You can use pd.get_dummies in EDA, but don't use it for modeling (instead, use OneHotEncoder, which works with Pipelines).
X = pd.get_dummies(tips_features)
X.head()
| total_bill | size | sex_Male | sex_Female | smoker_Yes | smoker_No | day_Thur | day_Fri | day_Sat | day_Sun | time_Lunch | time_Dinner | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 16.99 | 2 | 0 | 1 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | 1 |
| 1 | 10.34 | 3 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | 1 |
| 2 | 21.01 | 3 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | 1 |
| 3 | 23.68 | 2 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | 1 |
| 4 | 24.59 | 4 | 0 | 1 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | 1 |
Remember that under the hood, LinearRegression() creates a design matrix that has a column of all ones (for the intercept term). Let's add that column above for demonstration.
X['all_ones'] = 1
X.head()
| total_bill | size | sex_Male | sex_Female | smoker_Yes | smoker_No | day_Thur | day_Fri | day_Sat | day_Sun | time_Lunch | time_Dinner | all_ones | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 16.99 | 2 | 0 | 1 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | 1 | 1 |
| 1 | 10.34 | 3 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | 1 | 1 |
| 2 | 21.01 | 3 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | 1 | 1 |
| 3 | 23.68 | 2 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | 1 | 1 |
| 4 | 24.59 | 4 | 0 | 1 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | 1 | 1 |
Now, many of the above columns can be written as linear combinations of other columns!
'sex_Male' – its value is just 'all_ones' - 'sex_Female'.'smoker_Yes' – its value is just 'all_ones' - 'smoker_No'.'time_Lunch' – its value is just 'all_ones' - 'time_Dinner'.'day_Thur' – its value is just 'all_ones' - ('day_Fri' + 'day_Sat' + 'day_Sun').Note that if we get rid of the four redundant columns above, the rank of our design matrix – that is, the number of linearly independent columns it has – does not change (and so the "predictive power" of our features don't change either).
np.linalg.matrix_rank(X)
9
np.linalg.matrix_rank(X.drop(columns=['sex_Male', 'smoker_Yes', 'time_Lunch', 'day_Thur']))
9
However, without the redundant columns, there is only a single unique set of optimal parameters $w^*$, and the multicollinearity is no more.
Aside: Most one-hot encoding techniques (including OneHotEncoder) have an in-built drop argument, which allow you to specify that you'd like to drop one column per categorical feature.
pd.get_dummies(tips_features, drop_first=True)
| total_bill | size | sex_Female | smoker_No | day_Fri | day_Sat | day_Sun | time_Dinner | |
|---|---|---|---|---|---|---|---|---|
| 0 | 16.99 | 2 | 1 | 1 | 0 | 0 | 1 | 1 |
| 1 | 10.34 | 3 | 0 | 1 | 0 | 0 | 1 | 1 |
| 2 | 21.01 | 3 | 0 | 1 | 0 | 0 | 1 | 1 |
| 3 | 23.68 | 2 | 0 | 1 | 0 | 0 | 1 | 1 |
| 4 | 24.59 | 4 | 1 | 1 | 0 | 0 | 1 | 1 |
| ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 239 | 29.03 | 3 | 0 | 1 | 0 | 1 | 0 | 1 |
| 240 | 27.18 | 2 | 1 | 0 | 0 | 1 | 0 | 1 |
| 241 | 22.67 | 2 | 0 | 0 | 0 | 1 | 0 | 1 |
| 242 | 17.82 | 2 | 0 | 1 | 0 | 1 | 0 | 1 |
| 243 | 18.78 | 2 | 1 | 1 | 0 | 0 | 0 | 1 |
244 rows × 8 columns
from sklearn.preprocessing import OneHotEncoder
ohe = OneHotEncoder(drop='first')
ohe.fit_transform(tips_features[['sex', 'smoker', 'day', 'time']]).toarray()
array([[0., 0., 0., 1., 0., 0.],
[1., 0., 0., 1., 0., 0.],
[1., 0., 0., 1., 0., 0.],
...,
[1., 1., 1., 0., 0., 0.],
[1., 0., 1., 0., 0., 0.],
[0., 0., 0., 0., 1., 0.]])
The above array only has $(2-1) + (2-1) + (4-1) + (2-1) = 6$ columns, rather than $2 + 2 + 4 + 2 = 10$, since we dropped 1 per categorical column in tips_features.