import pandas as pd
import numpy as np
import os
import plotly.express as px
import plotly.graph_objects as go
pd.options.plotting.backend = 'plotly'
TEMPLATE = 'seaborn'
sklearn.
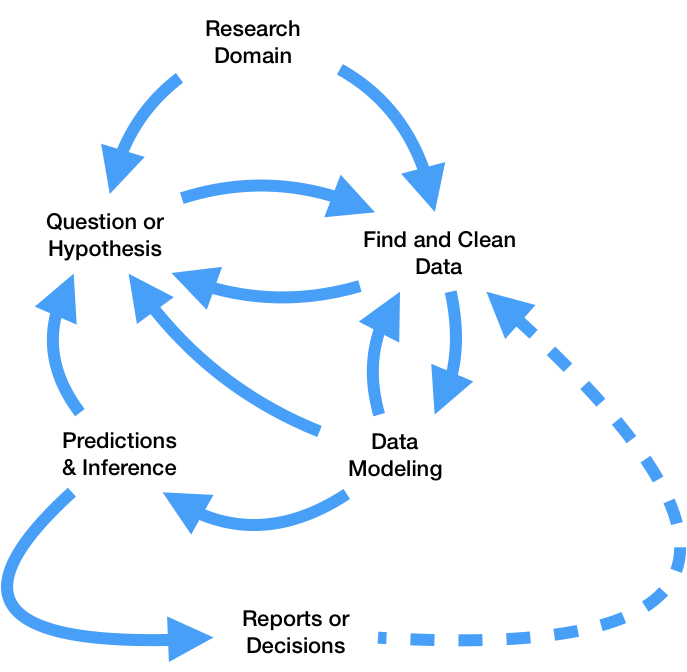
So far this quarter, we've learned how to:
pandas and regular expressions.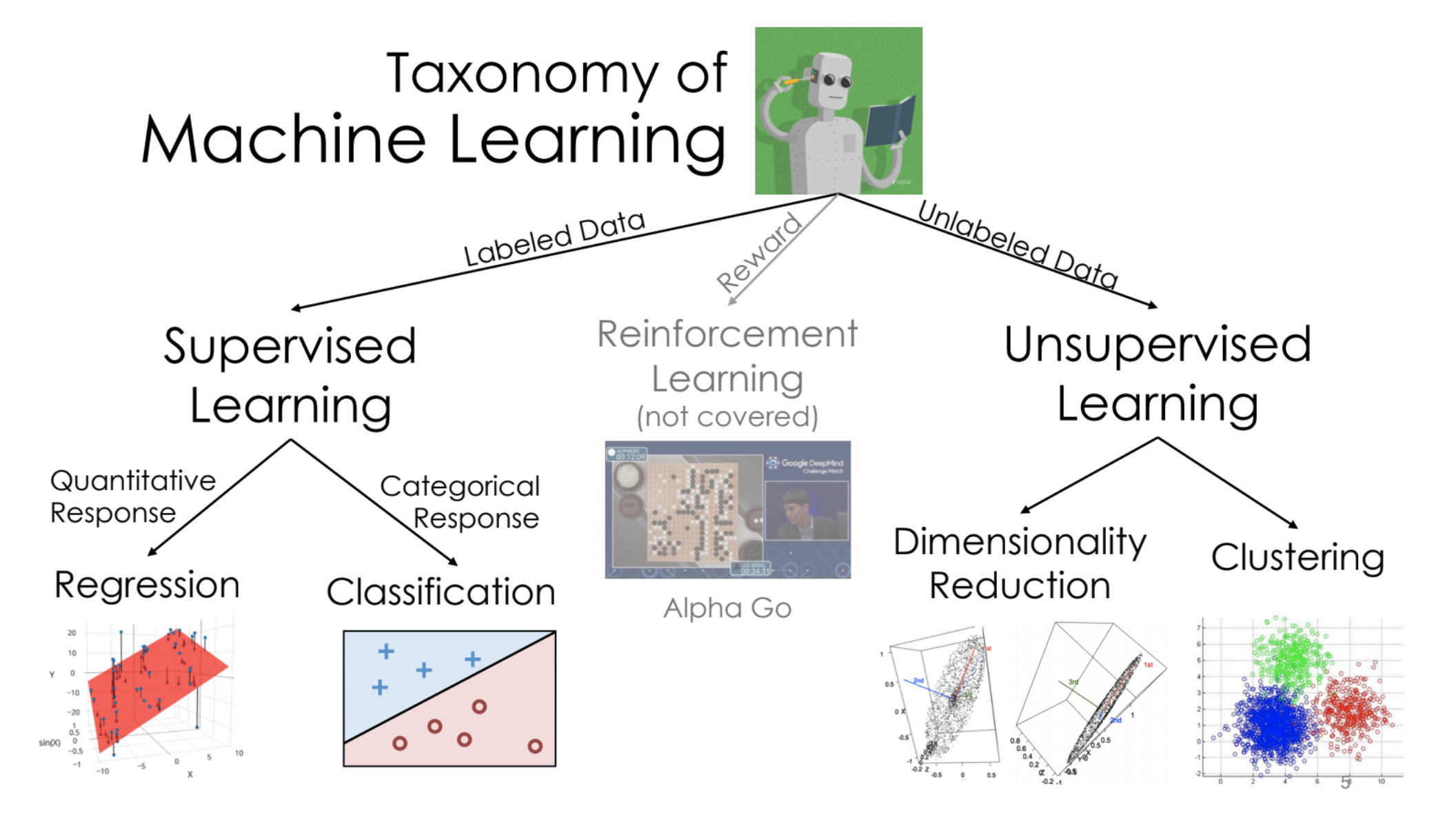
What features does the dataset contain?
# The dataset is built into plotly (and seaborn)!
tips = px.data.tips()
tips
| total_bill | tip | sex | smoker | day | time | size | |
|---|---|---|---|---|---|---|---|
| 0 | 16.99 | 1.01 | Female | No | Sun | Dinner | 2 |
| 1 | 10.34 | 1.66 | Male | No | Sun | Dinner | 3 |
| 2 | 21.01 | 3.50 | Male | No | Sun | Dinner | 3 |
| 3 | 23.68 | 3.31 | Male | No | Sun | Dinner | 2 |
| 4 | 24.59 | 3.61 | Female | No | Sun | Dinner | 4 |
| ... | ... | ... | ... | ... | ... | ... | ... |
| 239 | 29.03 | 5.92 | Male | No | Sat | Dinner | 3 |
| 240 | 27.18 | 2.00 | Female | Yes | Sat | Dinner | 2 |
| 241 | 22.67 | 2.00 | Male | Yes | Sat | Dinner | 2 |
| 242 | 17.82 | 1.75 | Male | No | Sat | Dinner | 2 |
| 243 | 18.78 | 3.00 | Female | No | Thur | Dinner | 2 |
244 rows × 7 columns
'total_bill'.'total_bill' and 'tip', as well as the distributions of both columns individually.tips.plot(kind='scatter',
x='total_bill', y='tip',
title='Tip vs. Total Bill',
template=TEMPLATE)
tips.plot(kind='hist',
x='total_bill',
title='Distribution of Total Bill',
nbins=50,
template=TEMPLATE)
tips.plot(kind='hist',
x='tip',
title='Distribution of Tip',
nbins=50,
template=TEMPLATE)
'total_bill' |
'tip' |
|---|---|
| Right skewed | Right skewed |
| Mean around $20 | Mean around $3 |
| Mode around $16 | Possibly bimodal at \$2 and \$3? |
| No particularly large bills | Large outliers? |
 George Box
George Box"Since all models are wrong the scientist cannot obtain a "correct" one by excessive elaboration. On the contrary following William of Occam he should seek an economical description of natural phenomena. Just as the ability to devise simple but evocative models is the signature of the great scientist so overelaboration and overparameterization is often the mark of mediocrity."
"Since all models are wrong the scientist must be alert to what is importantly wrong. It is inappropriate to be concerned about mice when there are tigers abroad."
Let's suppose we choose squared loss, meaning that $h^* = \text{mean}(y)$.
mean_tip = tips['tip'].mean()
mean_tip
2.99827868852459
Let's visualize this prediction.
# Unfortunately, the code to visualize a scatter plot and a line
# in plotly is not all that concise.
fig = go.Figure()
fig.add_trace(go.Scatter(
x=tips['total_bill'],
y=tips['tip'],
mode='markers',
name='Original Data')
)
fig.add_trace(go.Scatter(
x=[0, 60],
y=[mean_tip, mean_tip],
mode='lines',
name='Constant Prediction (Mean)'
))
fig.update_layout(showlegend=True, title='Tip vs. Total Bill',
xaxis_title='Total Bill', yaxis_title='Tip',
template=TEMPLATE)
fig.update_xaxes(range=[0, 60])
Note that to make predictions, this model ignores total bill (and all other features), and predicts the same tip for all tables.
np.mean((tips['tip'] - mean_tip) ** 2)
1.9066085124966412
# The same! A fact from 40A.
np.var(tips['tip'])
1.9066085124966412
Since we'll compute the RMSE for our future models too, we'll define a function that can compute it for us.
def rmse(actual, pred):
return np.sqrt(np.mean((actual - pred) ** 2))
Let's compute the RMSE of our constant tip's predictions, and store it in a dictionary that we can refer to later on.
rmse(tips['tip'], mean_tip)
1.3807999538298954
rmse_dict = {}
rmse_dict['constant tip amount'] = rmse(tips['tip'], mean_tip)
rmse_dict
{'constant tip amount': 1.3807999538298954}
Key idea: Since the mean minimizes RMSE for the constant model, it is impossible to change the mean_tip argument above to another number and yield a lower RMSE.
'total_bill'.tips.head()
| total_bill | tip | sex | smoker | day | time | size | |
|---|---|---|---|---|---|---|---|
| 0 | 16.99 | 1.01 | Female | No | Sun | Dinner | 2 |
| 1 | 10.34 | 1.66 | Male | No | Sun | Dinner | 3 |
| 2 | 21.01 | 3.50 | Male | No | Sun | Dinner | 3 |
| 3 | 23.68 | 3.31 | Male | No | Sun | Dinner | 2 |
| 4 | 24.59 | 3.61 | Female | No | Sun | Dinner | 4 |
A simple linear regression model is a linear model with a single feature, as we have here. For any total bill $x_i$, the predicted tip $H(x_i)$ is given by
$$H(x_i) = w_0 + w_1x_i$$sklearn¶sklearn¶
sklearn (scikit-learn) implements many common steps in the feature and model creation pipeline.numpy arrays, and to an extent, pandas DataFrames.LinearRegression class¶sklearn comes with several subpackages, including linear_model and tree, each of which contains several classes of models.LinearRegression class from linear_model.from sklearn.linear_model import LinearRegression
LinearRegression fits a linear model with coefficients w = (w1, …, wp) to minimize the residual sum of squares between the observed targets in the dataset, and the targets predicted by the linear approximation.
LinearRegression minimizes mean squared error by default! (Per the documentation, it also includes an intercept term by default.)LinearRegression?
First, we must instantiate a LinearRegression object and fit it. By calling fit, we are saying "minimize mean squared error on this dataset and find $w^*$."
model = LinearRegression()
# Note that there are two arguments to fit – X and y!
# (It is not necessary to write X= and y=)
model.fit(X=tips[['total_bill']], y=tips['tip'])
LinearRegression()
After fitting, we can access $w^*$ – that is, the best slope and intercept.
model.intercept_, model.coef_
(0.9202696135546731, array([0.10502452]))
These coefficients tell us that the "best way" (according to squared loss) to make tip predictions using a linear model is using:
$$\text{predicted tip} = 0.92 + 0.105 \cdot \text{total bill}$$This model assumes people tip by:
Let's visualize this model, along with our previous model.
fig.add_trace(go.Scatter(
x=[0, 60],
y=model.predict([[0], [60]]),
mode='lines',
name='Linear: Total Bill Only'
))
/Users/larry/opt/anaconda3/envs/dsc80/lib/python3.8/site-packages/sklearn/base.py:441: UserWarning: X does not have valid feature names, but LinearRegression was fitted with feature names
Visually, our linear model seems to be a better fit for our dataset than our constant model.
Fit LinearRegression objects also have a predict method, which can be used to predict tips for any total bill, new or old.
model.predict([[15]])
/Users/larry/opt/anaconda3/envs/dsc80/lib/python3.8/site-packages/sklearn/base.py:441: UserWarning: X does not have valid feature names, but LinearRegression was fitted with feature names
array([2.49563737])
# The input to model.predict **must** be a 2D list/array.
model.predict([[15],
[4],
[100]])
/Users/larry/opt/anaconda3/envs/dsc80/lib/python3.8/site-packages/sklearn/base.py:441: UserWarning: X does not have valid feature names, but LinearRegression was fitted with feature names
array([ 2.49563737, 1.34036768, 11.42272135])
model.predict(np.array(
[15, 4, 100]
).reshape(-1, 1))
/Users/larry/opt/anaconda3/envs/dsc80/lib/python3.8/site-packages/sklearn/base.py:441: UserWarning: X does not have valid feature names, but LinearRegression was fitted with feature names
array([ 2.49563737, 1.34036768, 11.42272135])
If we want to compute the RMSE of our model on the training data, we need to find its predictions on every row in the training data, tips.
all_preds = model.predict(tips[['total_bill']])
rmse_dict['one feature: total bill'] = rmse(tips['tip'], all_preds)
rmse_dict
{'constant tip amount': 1.3807999538298954,
'one feature: total bill': 1.0178504025697377}
tips we haven't touched:tips.head()
| total_bill | tip | sex | smoker | day | time | size | |
|---|---|---|---|---|---|---|---|
| 0 | 16.99 | 1.01 | Female | No | Sun | Dinner | 2 |
| 1 | 10.34 | 1.66 | Male | No | Sun | Dinner | 3 |
| 2 | 21.01 | 3.50 | Male | No | Sun | Dinner | 3 |
| 3 | 23.68 | 3.31 | Male | No | Sun | Dinner | 2 |
| 4 | 24.59 | 3.61 | Female | No | Sun | Dinner | 4 |
To find the optimal parameters $w^*$, we will again use sklearn's LinearRegression class. The code is not all that different!
model_two = LinearRegression()
model_two.fit(X=tips[['total_bill', 'size']], y=tips['tip'])
LinearRegression()
model_two.intercept_, model_two.coef_
(0.6689447408125031, array([0.09271334, 0.19259779]))
model_two.predict([[25, 4]])
/Users/larry/opt/anaconda3/envs/dsc80/lib/python3.8/site-packages/sklearn/base.py:441: UserWarning: X does not have valid feature names, but LinearRegression was fitted with feature names
array([3.75716934])
What does this model look like?
Here, we must draw a 3D scatter plot and plane, with one axis for total bill, one axis for table size, and one axis for tip. The code below does this.
XX, YY = np.mgrid[0:50:2, 0:8:1]
Z = model_two.intercept_ + model_two.coef_[0] * XX + model_two.coef_[1] * YY
plane = go.Surface(x=XX, y=YY, z=Z, colorscale='Oranges')
fig = go.Figure(data=[plane])
fig.add_trace(go.Scatter3d(x=tips['total_bill'],
y=tips['size'],
z=tips['tip'], mode='markers', marker = {'color': '#656DF1'}))
fig.update_layout(scene = dict(
xaxis_title='total bill',
yaxis_title='table size',
zaxis_title='tip'),
title='Tip vs. Total Bill and Table Size',
width=1000, height=800)
How does our two-feature linear model stack up to our single feature linear model and our constant model?
rmse_dict['two features'] = rmse(
tips['tip'], model_two.predict(tips[['total_bill', 'size']])
)
rmse_dict
{'constant tip amount': 1.3807999538298954,
'one feature: total bill': 1.0178504025697377,
'two features': 1.007256127114662}
LinearRegression class in sklearn.linear_model provides an implementation of least squares linear regression that works with multiple features.