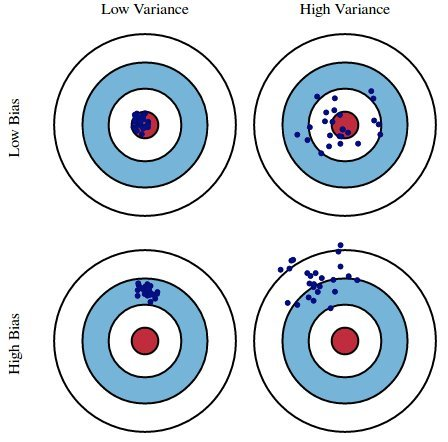
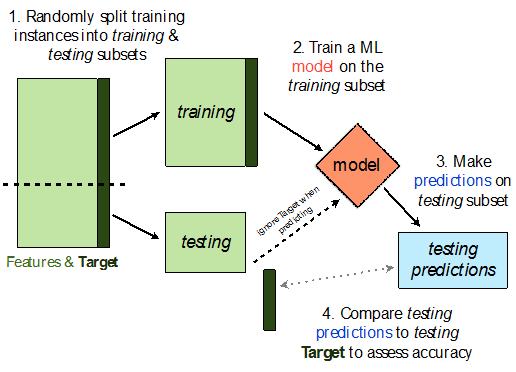
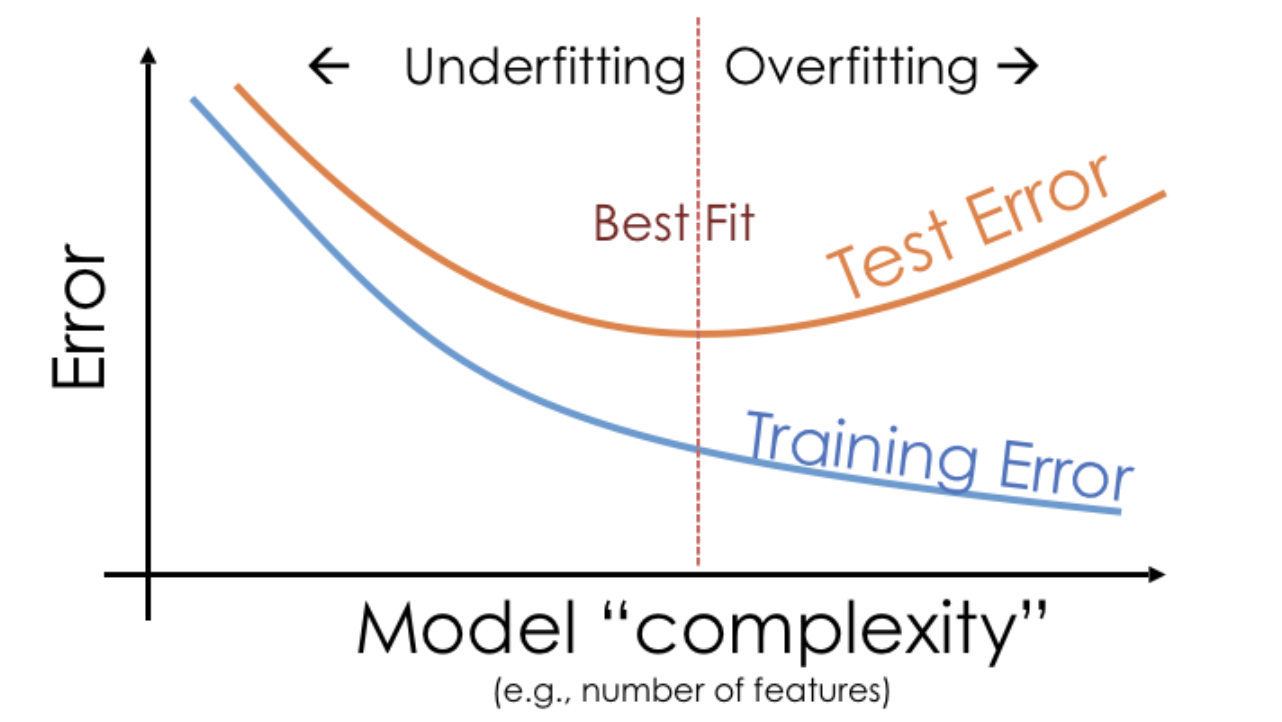
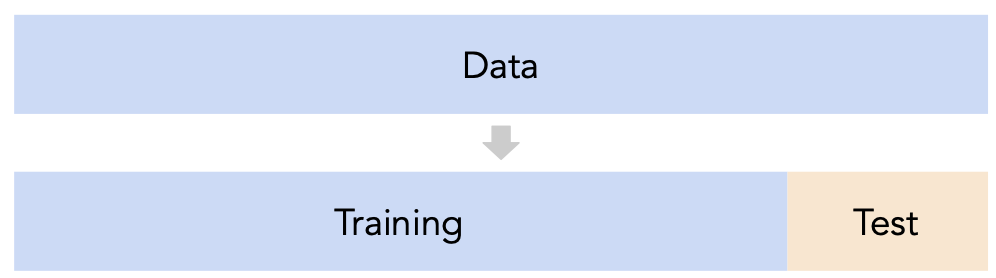
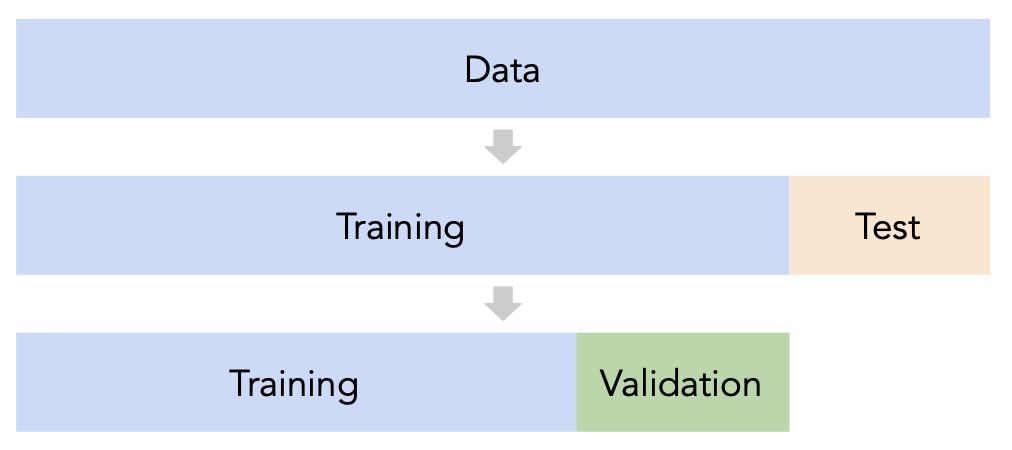
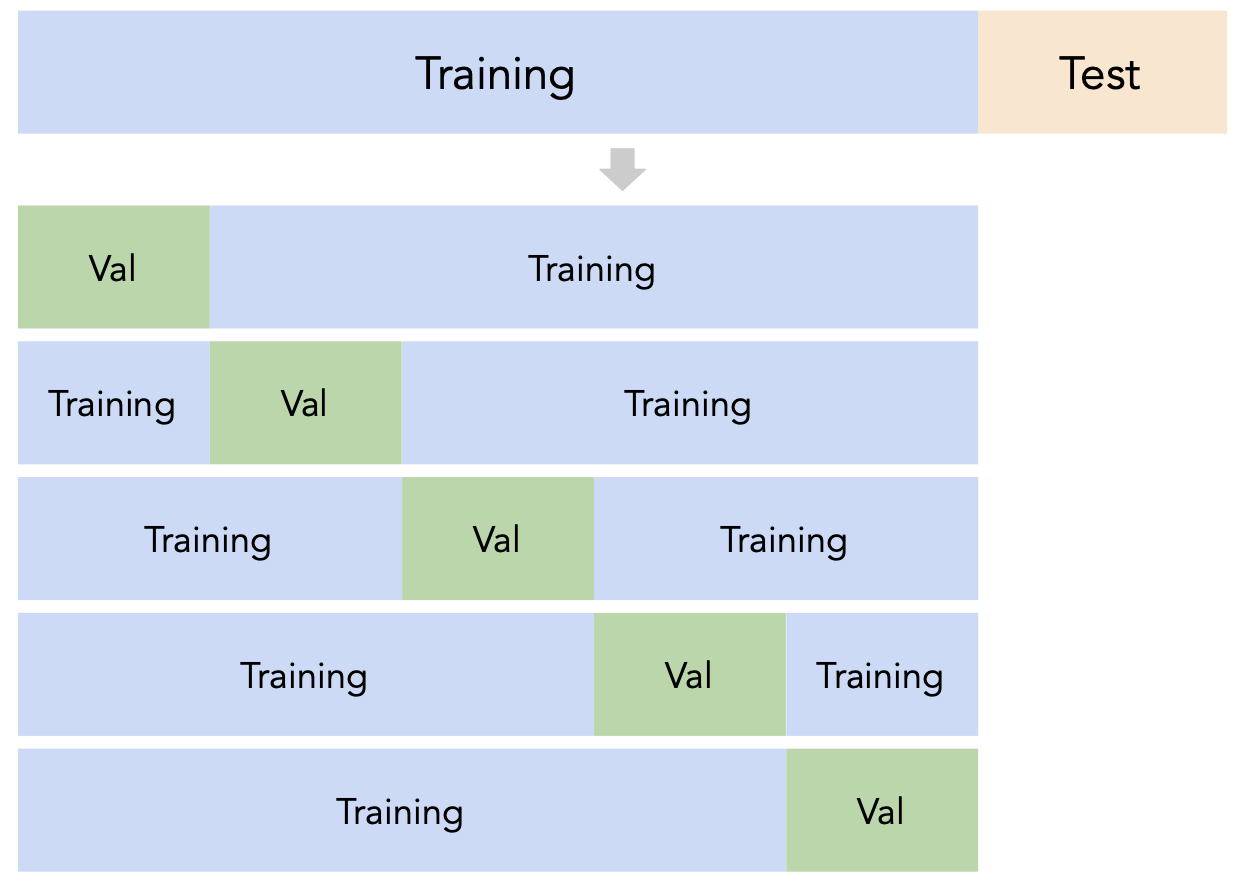
# A dictionary that maps names to Pipeline objects.
pipes = {
'total_bill only': Pipeline([
('trans', ColumnTransformer(
[('keep', FunctionTransformer(lambda x: x), ['total_bill'])],
remainder='drop')),
('lin-reg', LinearRegression())
]),
'total_bill + size': Pipeline([
('trans', ColumnTransformer(
[('keep', FunctionTransformer(lambda x: x), ['total_bill', 'size'])],
remainder='drop')),
('lin-reg', LinearRegression())
]),
'total_bill + size + OHE smoker': Pipeline([
('trans', ColumnTransformer(
[('keep', FunctionTransformer(lambda x: x), ['total_bill', 'size']),
('ohe', OneHotEncoder(), ['smoker'])],
remainder='drop')),
('lin-reg', LinearRegression())
]),
'total_bill + size + OHE all': Pipeline([
('trans', ColumnTransformer(
[('keep', FunctionTransformer(lambda x: x), ['total_bill', 'size']),
('ohe', OneHotEncoder(), ['smoker', 'sex', 'time', 'day'])],
remainder='drop')),
('lin-reg', LinearRegression())
]),
}