import pandas as pd
import numpy as np
import os
import seaborn as sns
import plotly.express as px
pd.options.plotting.backend = 'plotly'
Consider the penguins dataset from a few lectures ago.
penguins = sns.load_dataset('penguins').dropna()
penguins.head()
| species | island | bill_length_mm | bill_depth_mm | flipper_length_mm | body_mass_g | sex | |
|---|---|---|---|---|---|---|---|
| 0 | Adelie | Torgersen | 39.1 | 18.7 | 181.0 | 3750.0 | Male |
| 1 | Adelie | Torgersen | 39.5 | 17.4 | 186.0 | 3800.0 | Female |
| 2 | Adelie | Torgersen | 40.3 | 18.0 | 195.0 | 3250.0 | Female |
| 4 | Adelie | Torgersen | 36.7 | 19.3 | 193.0 | 3450.0 | Female |
| 5 | Adelie | Torgersen | 39.3 | 20.6 | 190.0 | 3650.0 | Male |
penguins.groupby('island')['bill_length_mm'].agg(['mean', 'count'])
| mean | count | |
|---|---|---|
| island | ||
| Biscoe | 45.248466 | 163 |
| Dream | 44.221951 | 123 |
| Torgersen | 39.038298 | 47 |
It appears that penguins on Torgersen Island have shorter bills on average than penguins on other islands.
Again, while you could do this with a for-loop (and you can use a for-loop for hypothesis tests in labs and projects), we'll use the faster size approach here.
Instead of using np.random.multinomial, which samples from a categorical distribution, we'll use np.random.choice, which samples from a known sequence of values.
# Draws two samples of size 47 from penguins['bill_length_mm'].
# Question: Why must we sample with replacement here (or, more specifically, in the next cell)?
np.random.choice(penguins['bill_length_mm'], size=(2, 47))
array([[39. , 43.8, 51.5, 52. , 37.6, 50.5, 41.6, 36.2, 51. , 37.6, 36.4,
37.2, 52. , 43.2, 39.2, 38.5, 40.3, 42.7, 50.2, 48.5, 49.5, 41.1,
45.2, 46.1, 35.5, 37.6, 52.5, 49.5, 39.6, 51.5, 50.4, 40.5, 44. ,
44.5, 45.1, 50.2, 45.2, 41.4, 48.8, 40.5, 50.6, 50. , 40.8, 39.2,
47.2, 51.3, 46.8],
[42.5, 44.5, 36.6, 45.1, 54.2, 49.4, 36.2, 37.9, 45.2, 49.3, 38.1,
36.4, 52.2, 39.7, 37.7, 44.9, 52. , 37. , 50.8, 45.1, 47.5, 42.3,
41.3, 46.8, 42.3, 41. , 55.8, 42.2, 38.8, 48.2, 34.6, 52.7, 39. ,
45.4, 41.1, 39.8, 36.4, 46.4, 40.9, 37.7, 37.2, 45.5, 42.8, 37.8,
46.4, 52. , 45. ]])
# Draws 100000 samples of size 47 from penguins['bill_length_mm'].
num_reps = 100_000
averages = np.random.choice(penguins['bill_length_mm'], size=(num_reps, 47)).mean(axis=1)
averages
array([43.9893617 , 43.31914894, 44.25319149, ..., 43.76382979,
43.5787234 , 43.04680851])
fig = px.histogram(pd.DataFrame(averages), x=0, nbins=50, histnorm='probability',
title='Empirical Distribution of the Average Bill Length in Samples of Size 47')
fig.add_vline(x=penguins.loc[penguins['island'] == 'Torgersen', 'bill_length_mm'].mean(), line_color='red')
It doesn't look like the average bill length of penguins on Torgersen Island came from the distribution of bill lengths of all penguins in our dataset.
There is a statistical tool you've learned about that would allow us to find the true probability distribution of the test statistic in this case. What is it?
penguins['bill_length_mm'] is roughly normal with mean penguins['bill_length_mm'].mean() and standard deviation penguins['bill_length_mm'].std(ddof=0) / np.sqrt(47).
"Standard" hypothesis testing helps us answer questions of the form:
I have a population distribution, and I have one sample. Does this sample look like it was drawn from the population?
It does not help us answer questions of the form:
I have two samples, but no information about any population distributions. Do these samples look like they were drawn from the same population?
That's where permutation testing comes in.
Note: For familiarity, we'll start with an example from DSC 10. This means we'll move quickly!
Let's start by loading in the data.
baby = pd.read_csv(os.path.join('data', 'baby.csv'))
baby.head()
| Birth Weight | Gestational Days | Maternal Age | Maternal Height | Maternal Pregnancy Weight | Maternal Smoker | |
|---|---|---|---|---|---|---|
| 0 | 120 | 284 | 27 | 62 | 100 | False |
| 1 | 113 | 282 | 33 | 64 | 135 | False |
| 2 | 128 | 279 | 28 | 64 | 115 | True |
| 3 | 108 | 282 | 23 | 67 | 125 | True |
| 4 | 136 | 286 | 25 | 62 | 93 | False |
We're only interested in the 'Birth Weight' and 'Maternal Smoker' columns.
baby = baby[['Maternal Smoker', 'Birth Weight']]
baby.head()
| Maternal Smoker | Birth Weight | |
|---|---|---|
| 0 | False | 120 |
| 1 | False | 113 |
| 2 | True | 128 |
| 3 | True | 108 |
| 4 | False | 136 |
Note that there are two samples:
How many babies are in each group? What is the average birth weight within each group?
baby.groupby('Maternal Smoker')['Birth Weight'].agg(['mean', 'count'])
| mean | count | |
|---|---|---|
| Maternal Smoker | ||
| False | 123.085315 | 715 |
| True | 113.819172 | 459 |
Note that 16 ounces are in 1 pound, so the above weights are ~7-8 pounds.
Below, we draw the distributions of both sets of birth weights.
px.histogram(baby, color='Maternal Smoker', histnorm='probability', marginal='box',
title="Birth Weight by Mother's Smoking Status", barmode='overlay', opacity=0.7)
There appears to be a difference, but can it be attributed to random chance?

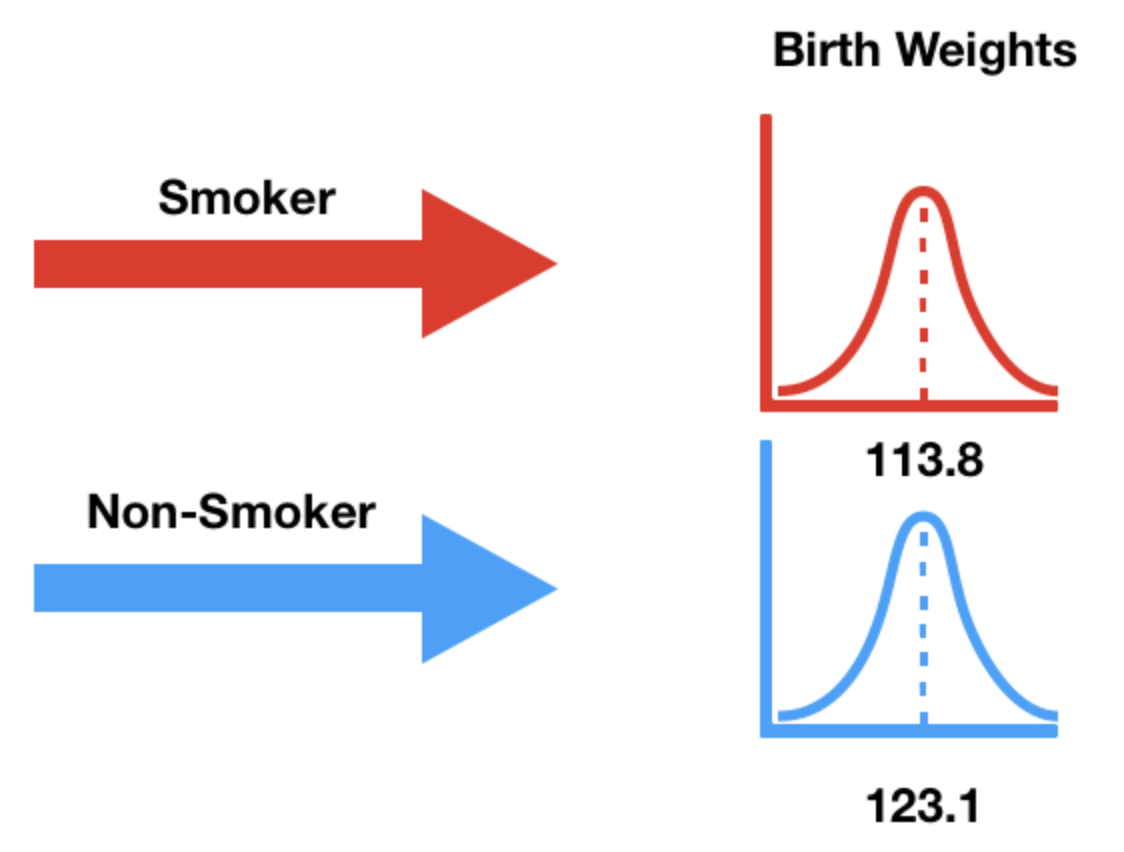
We need a test statistic that can measure how different two numerical distributions are.
px.histogram(baby, color='Maternal Smoker', histnorm='probability', marginal='box',
title="Birth Weight by Mother's Smoking Status", barmode='overlay', opacity=0.7)
Easiest solution: Difference in group means.
We'll choose our test statistic to be:
$$\text{mean weight of smokers' babies} - \text{mean weight of non-smokers' babies}$$We could also compute the non-smokers' mean minus the smokers' mean, too.
group_means = baby.groupby('Maternal Smoker')['Birth Weight'].mean()
group_means
Maternal Smoker False 123.085315 True 113.819172 Name: Birth Weight, dtype: float64
At first, you may think to use loc with group_means to compute the difference in group means.
group_means.loc[True] - group_means.loc[False]
-9.266142572024918
However, you can also use the diff method.
pd.Series([1, 2, -3]).diff()
0 NaN 1 1.0 2 -5.0 dtype: float64
group_means.diff()
Maternal Smoker False NaN True -9.266143 Name: Birth Weight, dtype: float64
group_means.diff()[-1]
-9.266142572024918
# If we wanted to do (non-smokers' mean - smokers' mean).
# Think about why this is the case (hint: it has to do with how the resulting DataFrame after grouping is sorted).
group_means[::-1].diff()[-1]
9.266142572024918
'Maternal Smoker' – has no effect on the birth weight.True or False to each baby.baby.head()
| Maternal Smoker | Birth Weight | |
|---|---|---|
| 0 | False | 120 |
| 1 | False | 113 |
| 2 | True | 128 |
| 3 | True | 108 |
| 4 | False | 136 |
True or False, while keeping the same number of Trues and Falses as we started with.'Maternal Smoker' column to values in the 'Birth Weight' column.np.random.permutation.df.sample, but it's more complicated.np.random.permutation(baby['Birth Weight'])
array([135, 94, 125, ..., 124, 121, 134])
with_shuffled = baby.assign(Shuffled_Weights=np.random.permutation(baby['Birth Weight']))
with_shuffled.head()
| Maternal Smoker | Birth Weight | Shuffled_Weights | |
|---|---|---|---|
| 0 | False | 120 | 114 |
| 1 | False | 113 | 139 |
| 2 | True | 128 | 129 |
| 3 | True | 108 | 133 |
| 4 | False | 136 | 114 |
'Birth Weights' and assigned them to the smokers' group, and the remaining 715 to the non-smokers' group.One benefit of shuffling 'Birth Weight' (instead of 'Maternal Smoker') is that grouping by 'Maternal Smoker' allows us to see all of the following information with a single call to groupby.
group_means = with_shuffled.groupby('Maternal Smoker').mean()
group_means
| Birth Weight | Shuffled_Weights | |
|---|---|---|
| Maternal Smoker | ||
| False | 123.085315 | 119.706294 |
| True | 113.819172 | 119.082789 |
Let's visualize both pairs of distributions – what do you notice?
for x in ['Birth Weight', 'Shuffled_Weights']:
fig = px.histogram(with_shuffled, x=x, color='Maternal Smoker', histnorm='probability', marginal='box',
title=f"Using the {x} column (difference in means = {group_means[x].diff().iloc[-1]})", barmode='overlay', opacity=0.7)
fig.show()
n_repetitions = 500
differences = []
for _ in range(n_repetitions):
# Step 1: Shuffle the weights and store them in a DataFrame.
with_shuffled = baby.assign(Shuffled_Weights=np.random.permutation(baby['Birth Weight']))
# Step 2: Compute the test statistic.
# Remember, alphabetically, False comes before True,
# so this computes True - False.
group_means = (
with_shuffled
.groupby('Maternal Smoker')
.mean()
.loc[:, 'Shuffled_Weights']
)
difference = group_means.diff().iloc[-1]
# Step 4: Store the result
differences.append(difference)
differences[:10]
[-0.1048036930389884, -0.24073921720980707, -0.3909837439249202, -0.469683257918561, 1.1758520346755716, 0.3530843883785053, -0.4017154958331446, -2.2082270670505864, -0.42675625028566344, -3.063189969072326]
We already computed the observed statistic earlier, but we compute it again below to keep all of our calculations together.
observed_difference = baby.groupby('Maternal Smoker')['Birth Weight'].mean().diff().iloc[-1]
observed_difference
-9.266142572024918
fig = px.histogram(pd.DataFrame(differences), x=0, nbins=50, histnorm='probability',
title='Empirical Distribution of the Mean Differences in Birth Weights (Smoker - Non-Smoker)')
fig.add_vline(x=observed_difference, line_color='red')
fig.update_layout(xaxis_range=[-15, 15])
couples_fp = os.path.join('data', 'married_couples.csv')
couples = pd.read_csv(couples_fp)
couples.head()
| hh_id | gender | mar_status | rel_rating | age | education | hh_income | empl_status | hh_internet | |
|---|---|---|---|---|---|---|---|---|---|
| 0 | 0 | 1 | 1 | 1 | 51 | 12 | 14 | 1 | 1 |
| 1 | 0 | 2 | 1 | 1 | 53 | 9 | 14 | 1 | 1 |
| 2 | 1 | 1 | 1 | 1 | 57 | 11 | 15 | 1 | 1 |
| 3 | 1 | 2 | 1 | 1 | 57 | 9 | 15 | 1 | 1 |
| 4 | 2 | 1 | 1 | 1 | 60 | 12 | 14 | 1 | 1 |
# What does this expression compute?
couples['hh_id'].value_counts().value_counts()
2 1033 1 2 Name: hh_id, dtype: int64
We won't use all of the columns in the DataFrame.
couples = couples[['mar_status', 'empl_status', 'gender', 'age']]
couples.head()
| mar_status | empl_status | gender | age | |
|---|---|---|---|---|
| 0 | 1 | 1 | 1 | 51 |
| 1 | 1 | 1 | 2 | 53 |
| 2 | 1 | 1 | 1 | 57 |
| 3 | 1 | 1 | 2 | 57 |
| 4 | 1 | 1 | 1 | 60 |
The numbers in the DataFrame correspond to the mappings below.
'mar_status': 1=married, 2=unmarried.'empl_status': enumerated in the list below.'gender': 1=male, 2=female.'age': person's age in years.couples.head()
| mar_status | empl_status | gender | age | |
|---|---|---|---|---|
| 0 | 1 | 1 | 1 | 51 |
| 1 | 1 | 1 | 2 | 53 |
| 2 | 1 | 1 | 1 | 57 |
| 3 | 1 | 1 | 2 | 57 |
| 4 | 1 | 1 | 1 | 60 |
empl = [
'Working as paid employee',
'Working, self-employed',
'Not working - on a temporary layoff from a job',
'Not working - looking for work',
'Not working - retired',
'Not working - disabled',
'Not working - other'
]
couples = couples.replace({
'mar_status': {1: 'married', 2: 'unmarried'},
'gender': {1: 'M', 2: 'F'},
'empl_status': {(k + 1): empl[k] for k in range(len(empl))}
})
couples.head()
| mar_status | empl_status | gender | age | |
|---|---|---|---|---|
| 0 | married | Working as paid employee | M | 51 |
| 1 | married | Working as paid employee | F | 53 |
| 2 | married | Working as paid employee | M | 57 |
| 3 | married | Working as paid employee | F | 57 |
| 4 | married | Working as paid employee | M | 60 |
couples dataset¶# For categorical columns, this shows the 10 most common values and their frequencies.
# For numerical columns, this shows the result of calling the .describe() method.
for col in couples:
if couples[col].dtype == 'object':
empr = couples[col].value_counts(normalize=True).to_frame().iloc[:10]
else:
empr = couples[col].describe().to_frame()
display(empr)
| mar_status | |
|---|---|
| married | 0.717602 |
| unmarried | 0.282398 |
| empl_status | |
|---|---|
| Working as paid employee | 0.605899 |
| Not working - other | 0.103965 |
| Working, self-employed | 0.098646 |
| Not working - looking for work | 0.067698 |
| Not working - disabled | 0.056576 |
| Not working - retired | 0.050774 |
| Not working - on a temporary layoff from a job | 0.016441 |
| gender | |
|---|---|
| M | 0.5 |
| F | 0.5 |
| age | |
|---|---|
| count | 2068.000000 |
| mean | 43.165377 |
| std | 11.906982 |
| min | 18.000000 |
| 25% | 33.000000 |
| 50% | 44.000000 |
| 75% | 53.000000 |
| max | 64.000000 |
Let's look at the distribution of age separately for married couples and unmarried couples.
px.histogram(couples, x='age', color='mar_status', histnorm='probability', marginal='box',
barmode='overlay', opacity=0.7)
How are these two distributions different? Why do you think there is a difference?
To answer these questions, let's compute the distribution of employment status conditional on household type (married vs. unmarried).
couples.sample(5).head()
| mar_status | empl_status | gender | age | |
|---|---|---|---|---|
| 1844 | unmarried | Working as paid employee | M | 57 |
| 1074 | married | Working as paid employee | M | 34 |
| 335 | married | Working as paid employee | F | 52 |
| 1542 | married | Working as paid employee | M | 35 |
| 840 | married | Working as paid employee | M | 33 |
# Note that this is a shortcut to picking a column for values and using aggfunc='count'.
empl_cnts = couples.pivot_table(index='empl_status', columns='mar_status', aggfunc='size')
empl_cnts
| mar_status | married | unmarried |
|---|---|---|
| empl_status | ||
| Not working - disabled | 72 | 45 |
| Not working - looking for work | 71 | 69 |
| Not working - on a temporary layoff from a job | 21 | 13 |
| Not working - other | 182 | 33 |
| Not working - retired | 94 | 11 |
| Working as paid employee | 906 | 347 |
| Working, self-employed | 138 | 66 |
Since there are a different number of married and unmarried couples in the dataset, we can't compare the numbers above directly. We need to convert counts to proportions, separately for married and unmarried couples.
empl_cnts.sum()
mar_status married 1484 unmarried 584 dtype: int64
cond_distr = empl_cnts / empl_cnts.sum()
cond_distr
| mar_status | married | unmarried |
|---|---|---|
| empl_status | ||
| Not working - disabled | 0.048518 | 0.077055 |
| Not working - looking for work | 0.047844 | 0.118151 |
| Not working - on a temporary layoff from a job | 0.014151 | 0.022260 |
| Not working - other | 0.122642 | 0.056507 |
| Not working - retired | 0.063342 | 0.018836 |
| Working as paid employee | 0.610512 | 0.594178 |
| Working, self-employed | 0.092992 | 0.113014 |
Both of the columns above sum to 1.
Are the distributions of employment status for married people and for unmarried people who live with their partners different?
Is this difference just due to noise?
cond_distr.plot(kind='barh', title='Distribution of Employment Status, Conditional on Household Type', barmode='group')
Null Hypothesis: In the US, the distribution of employment status among those who are married is the same as among those who are unmarried and live with their partners. The difference between the two observed samples is due to chance.
Alternative Hypothesis: In the US, the distributions of employment status of the two groups are different.
What is a good test statistic in this case?
Hint: What kind of distributions are we comparing?
cond_distr
| mar_status | married | unmarried |
|---|---|---|
| empl_status | ||
| Not working - disabled | 0.048518 | 0.077055 |
| Not working - looking for work | 0.047844 | 0.118151 |
| Not working - on a temporary layoff from a job | 0.014151 | 0.022260 |
| Not working - other | 0.122642 | 0.056507 |
| Not working - retired | 0.063342 | 0.018836 |
| Working as paid employee | 0.610512 | 0.594178 |
| Working, self-employed | 0.092992 | 0.113014 |
Let's first compute the observed TVD, using our new knowledge of the diff method.
cond_distr.diff(axis=1).iloc[:, -1].abs().sum() / 2
0.1269754089281099
Since we'll need to calculate the TVD repeatedly, let's define a function that computes it.
def tvd_of_groups(df, groups, cats):
'''groups: the binary column (e.g. married vs. unmarried).
cats: the categorical column (e.g. employment status).
'''
cnts = df.pivot_table(index=cats, columns=groups, aggfunc='size')
# Normalize each column.
distr = cnts / cnts.sum()
# Compute and return the TVD.
return distr.diff(axis=1).iloc[:, -1].abs().sum() / 2
# Same result as above.
observed_tvd = tvd_of_groups(couples, groups='mar_status', cats='empl_status')
observed_tvd
0.1269754089281099
couples.head()
| mar_status | empl_status | gender | age | |
|---|---|---|---|---|
| 0 | married | Working as paid employee | M | 51 |
| 1 | married | Working as paid employee | F | 53 |
| 2 | married | Working as paid employee | M | 57 |
| 3 | married | Working as paid employee | F | 57 |
| 4 | married | Working as paid employee | M | 60 |
Here, we'll shuffle marital statuses, though remember, we could shuffle employment statuses too.
couples.assign(shuffled_mar=np.random.permutation(couples['mar_status']))
| mar_status | empl_status | gender | age | shuffled_mar | |
|---|---|---|---|---|---|
| 0 | married | Working as paid employee | M | 51 | married |
| 1 | married | Working as paid employee | F | 53 | married |
| 2 | married | Working as paid employee | M | 57 | unmarried |
| 3 | married | Working as paid employee | F | 57 | unmarried |
| 4 | married | Working as paid employee | M | 60 | married |
| ... | ... | ... | ... | ... | ... |
| 2063 | unmarried | Working as paid employee | F | 42 | married |
| 2064 | unmarried | Working as paid employee | M | 60 | married |
| 2065 | unmarried | Working as paid employee | F | 53 | unmarried |
| 2066 | unmarried | Working as paid employee | M | 44 | married |
| 2067 | unmarried | Working as paid employee | F | 42 | married |
2068 rows × 5 columns
Let's do this repeatedly.
N = 1000
tvds = []
for _ in range(N):
# Shuffle marital statuses.
with_shuffled = couples.assign(shuffled_mar=np.random.permutation(couples['mar_status']))
# Compute and store the TVD.
tvd = tvd_of_groups(with_shuffled, groups='shuffled_mar', cats='empl_status')
tvds.append(tvd)
Notice that by defining a function that computes our test statistic, our simulation code is much cleaner.
fig = px.histogram(pd.DataFrame(tvds), x=0, nbins=50, histnorm='probability',
title='Empirical Distribution of the TVD')
fig.add_vline(x=observed_tvd, line_color='red')
fig.add_annotation(text=f'<span style="color:red">Observed TVD = {round(observed_tvd, 2)}</span>',
x=1.15 * observed_tvd, showarrow=False, y=0.055)
fig.update_layout(xaxis_range=[0, 0.2])
p_95 = np.percentile(tvds, 95)
fig.add_vline(x=p_95, line_color='purple')
annot_text = f'<span style="color:purple">The 95th percentile of our<br>empirical distribution is {round(p_95, 2)}.<br><br>'
annot_text += 'If our observed statistic is to the<br>right of this point, we will reject the null<br>at a 5% <b>significance level</b>.</span>'
fig.add_annotation(text=annot_text, x=1.5 * np.percentile(tvds, 95), showarrow=False, y=0.05)
We reject the null hypothesis that married/unmarried households have similar employment makeups.
We can't say anything about why the employment makeups are different, though!
In the definition of the TVD, we divide the sum of the absolute differences in proportions between the two distributions by 2.
def tvd(a, b):
return np.sum(np.abs(a - b)) / 2
Question: If we divided by 200 instead of 2, would we still reject the null hypothesis?
Wrap up hypothesis and permutation testing. Revisit missing values.