from dsc80_utils import *
Announcements 📣¶
- Lab 8 due tomorrow.
- Final Project Checkpoint 2 due Tuesday.
Agenda¶
- Grid search
- Random forests
- Modeling with text features
- Classifier evaluation
Generalization¶
Question 🤔 (Answer at dsc80.com/q)
Code: 10cv
- Suppose you have a training dataset with 1000 rows.
- You want to decide between 20 hyperparameters for a particular model.
- To do so, you perform 10-fold cross-validation.
- How many times is the first row in the training dataset (
X.iloc[0]) used for training a model?
Summary: Generalization¶
- Split the data into two sets: training and test.
- Use only the training data when designing, training, and tuning the model.
- Use $k$-fold cross-validation to choose hyperparameters and estimate the model's ability to generalize.
- Do not ❌ look at the test data in this step!
- Commit to your final model and train it using the entire training set.
- Test the data using the test data. If the performance (e.g. RMSE) is not acceptable, return to step 2.
- Finally, train on all available data and ship the model to production! 🛳
🚨 This is the process you should always use! 🚨
Decision trees 🌲¶
Decision trees can be used for both regression and classification. We'll start by using them for classification.
Example: Should I get groceries?¶
Decision trees make classifications by answering a series of yes/no questions.
Should I go to Trader Joe's to buy groceries today?
Internal nodes of trees involve questions; leaf nodes make predictions $H(x)$.
Example: Predicting diabetes¶
diabetes = pd.read_csv(Path('data') / 'diabetes.csv')
display_df(diabetes, cols=9)
| Pregnancies | Glucose | BloodPressure | SkinThickness | Insulin | BMI | DiabetesPedigreeFunction | Age | Outcome | |
|---|---|---|---|---|---|---|---|---|---|
| 0 | 6 | 148 | 72 | 35 | 0 | 33.6 | 0.63 | 50 | 1 |
| 1 | 1 | 85 | 66 | 29 | 0 | 26.6 | 0.35 | 31 | 0 |
| 2 | 8 | 183 | 64 | 0 | 0 | 23.3 | 0.67 | 32 | 1 |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 765 | 5 | 121 | 72 | 23 | 112 | 26.2 | 0.24 | 30 | 0 |
| 766 | 1 | 126 | 60 | 0 | 0 | 30.1 | 0.35 | 47 | 1 |
| 767 | 1 | 93 | 70 | 31 | 0 | 30.4 | 0.32 | 23 | 0 |
768 rows × 9 columns
# 0 means no diabetes, 1 means yes diabetes.
diabetes['Outcome'].value_counts()
Outcome 0 500 1 268 Name: count, dtype: int64
'Glucose'is measured in mg/dL (milligrams per deciliter).
'BMI'is calculated as $\text{BMI} = \frac{\text{weight (kg)}}{\left[ \text{height (m)} \right]^2}$.
- Let's use
'Glucose'and'BMI'to predict whether or not a patient has diabetes ('Outcome').
Exploring the dataset¶
First, a train-test split:
from sklearn.model_selection import train_test_split
X_train, X_test, y_train, y_test = (
train_test_split(diabetes[['Glucose', 'BMI']], diabetes['Outcome'], random_state=1)
)
Class 0 (orange) is "no diabetes" and class 1 (blue) is "diabetes".
fig = (
X_train.assign(Outcome=y_train.astype(str))
.plot(kind='scatter', x='Glucose', y='BMI', color='Outcome',
color_discrete_map={'0': 'orange', '1': 'blue'},
title='Relationship between Glucose, BMI, and Diabetes')
)
fig
Building a decision tree¶
Let's build a decision tree and interpret the results.
The relevant class is DecisionTreeClassifier, from sklearn.tree.
from sklearn.tree import DecisionTreeClassifier
Note that we fit it the same way we fit earlier estimators.
You may wonder what max_depth and criterion do – more on this soon!
dt = DecisionTreeClassifier(max_depth=2, criterion='entropy')
dt.fit(X_train, y_train)
DecisionTreeClassifier(criterion='entropy', max_depth=2)In a Jupyter environment, please rerun this cell to show the HTML representation or trust the notebook.
On GitHub, the HTML representation is unable to render, please try loading this page with nbviewer.org.
DecisionTreeClassifier(criterion='entropy', max_depth=2)
Visualizing decision trees¶
Our fit decision tree is like a "flowchart", made up of a series of questions.
As before, orange is "no diabetes" and blue is "diabetes".
from sklearn.tree import plot_tree
plt.figure(figsize=(15, 5))
plot_tree(dt, feature_names=X_train.columns, class_names=['no db', 'yes db'],
filled=True, fontsize=15, impurity=False);
- To classify a new data point, we start at the top and answer the first question (i.e. "Glucose <= 129.5").
- If the answer is "Yes", we move to the left branch, otherwise we move to the right branch.
- We repeat this process until we end up at a leaf node, at which point we predict the most common class in that node.
- Note that each node has a
valueattribute, which describes the number of training individuals of each class that fell in that node.
- Note that each node has a
# Note that the left node at depth 2 has a `value` of [304, 78].
y_train[X_train.query('Glucose <= 129.5').index].value_counts()
Outcome 0 304 1 78 Name: count, dtype: int64
Evaluating classifiers¶
The most common evaluation metric in classification is accuracy:
$$\text{accuracy} = \frac{\text{\# data points classified correctly}}{\text{\# data points}}$$
(dt.predict(X_train) == y_train).mean()
np.float64(0.765625)
The score method of a classifier computes accuracy by default (just like the score method of a regressor computes $R^2$ by default). We want our classifiers to have high accuracy.
# Training accuracy – same number as above
dt.score(X_train, y_train)
0.765625
# Testing accuracy
dt.score(X_test, y_test)
0.7760416666666666
Reflection¶
- Decision trees are easily interpretable: it is clear how they make their predictions.
- They work with categorical data without needing to use one hot encoding.
- They also can be used in multi-class classification problems, e.g. when there are more than 2 possible outcomes.
- The decision boundary of a decision tree can be arbitrarily complicated.
- How are decision trees trained?
How are decision trees trained?¶
Pseudocode:
def make_tree(X, y):
if all points in y have the same label C:
return Leaf(C)
f = best splitting feature # e.g. Glucose or BMI
v = best splitting value # e.g. 129.5
X_left, y_left = X, y where (X[f] <= v)
X_right, y_right = X, y where (X[f] > v)
left = make_tree(X_left, y_left)
right = make_tree(X_right, y_right)
return Node(f, v, left, right)
make_tree(X_train, y_train)
How do we measure the quality of a split?¶
Our pseudocode for training a decision tree relies on finding the best way to "split" a node – that is, the best question to ask to help us narrow down which class to predict.
Intuition: Suppose the distribution within a node looks like this (colors represent classes):
Split A:
- "Yes": 🟠🟠🟠🔵🔵🔵
- "No": 🟠🟠🟠🔵🔵🔵🔵
Split B:
- "Yes": 🔵🔵🔵🔵🔵🔵
- "No": 🔵🟠🟠🟠🟠🟠🟠
Which split is "better"?
Split B, because there is "less uncertainty" in the resulting nodes in Split B than there is in Split A. Let's try and quantify this!
Entropy¶
- For each class $C$ within a node, define $p_C$ as the proportion of points with that class.
- For example, the two classes may be "yes diabetes" and "no diabetes".
- The surprise of drawing a point from the node at random and having it be class $C$ is:
$$ - \log_2 p_C $$
- The entropy of a node is the average surprise over all classes:
\begin{align} \text{entropy} &= - \sum_C p_C \log_2 p_C \end{align}
- The entropy of 🟠🟠🟠🟠🟠🟠🟠🟠 is $ -1 \log_2(1) = 0 $.
- The entropy of 🟠🟠🟠🟠🔵🔵🔵🔵 is $ -0.5 \log_2(0.5) - 0.5 \log_2(0.5) = 1$.
- The entropy of 🟠🔵🟢🟡🟣 is $ - \log_2 \frac{1}{5} = \log_2(5) $
- In general, if a node has $n$ points, all with different labels, the entropy of the node is $ \log_2(n) $.
Example entropy calculation¶
Suppose we have:
Split A:
- "Yes": 🟠🟠🟠🟠🟠🟠🔵
- "No": 🟠🟠🟠🟠🟠🟠🔵🔵🔵🔵🔵
Split B:
- "Yes": 🟠🟠🟠🟠🟠🟠🔵🔵🔵
- "No": 🟠🟠🟠🟠🟠🟠🔵🔵🔵
We choose the split that has the lowest weighted entropy, that is:
$$\text{entropy of split} = \frac{\# \text{Yes}}{\# \text{Yes} + \# \text{No}} \cdot \text{entropy(Yes)} + \frac{\# \text{No}}{\# \text{Yes} + \# \text{No}} \cdot \text{entropy(No)}$$
def entropy(node):
props = pd.Series(list(node)).value_counts(normalize=True)
return -sum(props * np.log2(props))
def weighted_entropy(yes_node, no_node):
yes_entropy = entropy(yes_node)
no_entropy = entropy(no_node)
yes_weight = len(yes_node) / (len(yes_node) + len(no_node))
return yes_weight * yes_entropy + (1 - yes_weight) * no_entropy
# Split A:
weighted_entropy("🟠🟠🟠🟠🟠🟠🔵", "🟠🟠🟠🟠🟠🟠🔵🔵🔵🔵🔵")
0.8375578764623786
# Split B:
weighted_entropy("🟠🟠🟠🟠🟠🟠🔵🔵🔵", "🟠🟠🟠🟠🟠🟠🔵🔵🔵")
0.9182958340544896
Split A has the lower weighted entropy, so we'll use it.
Understanding entropy¶
plt.figure(figsize=(15, 5))
plot_tree(dt, feature_names=X_train.columns, class_names=['no db', 'yes db'],
filled=True, fontsize=15, impurity=True);
We can recreate the entropy calculations above! Note that the default DecisionTreeClassifier uncertaintly metric isn't entropy; it used entropy because we set criterion='entropy' when defining dt. (The default metric, Gini impurity, is perfectly fine too!)
# The first node at depth 2 has an entropy of 0.73,
# both told to us above and verified here!
entropy([0] * 304 + [1] * 78)
0.7302263747422792
Tree depth¶
Decision trees are trained by recursively picking the best split until:
- all "leaf nodes" only contain training examples from a single class (good), or
- it is impossible to split leaf nodes any further (not good).
By default, there is no "maximum depth" for a decision tree. As such, without restriction, decision trees tend to be very deep.
dt_no_max = DecisionTreeClassifier()
dt_no_max.fit(X_train, y_train)
DecisionTreeClassifier()In a Jupyter environment, please rerun this cell to show the HTML representation or trust the notebook.
On GitHub, the HTML representation is unable to render, please try loading this page with nbviewer.org.
DecisionTreeClassifier()
A decision tree fit on our training data has a depth of around 20! (It is so deep that tree.plot_tree errors when trying to plot it.)
dt_no_max.tree_.max_depth
21
At first, this tree seems "better" than our tree of depth 2, since its training accuracy is much much higher:
dt_no_max.score(X_train, y_train)
0.9913194444444444
# Depth 2 tree.
dt.score(X_train, y_train)
0.765625
But recall, we truly care about test set performance, and this decision tree has worse accuracy on the test set than our depth 2 tree.
dt_no_max.score(X_test, y_test)
0.7135416666666666
# Depth 2 tree.
dt.score(X_test, y_test)
0.7760416666666666
Decision trees and overfitting¶
- Decision trees have a tendency to overfit. Why is that?
- Unlike linear classification techniques (like logistic regression), decision trees are non-linear.
- They are also "non-parametric" – there are no $w^*$s to learn.
- While being trained, decision trees ask enough questions to effectively memorize the correct response values in the training set. However, the relationships they learn are often overfit to the noise in the training set, and don't generalize well.
fig
- A decision tree whose depth is not restricted will achieve 100% accuracy on any training set, as long as there are no "overlapping values" in the training set.
- Two values overlap when they have the same features $x$ but different response values $y$ (e.g. if two patients have the same glucose levels and BMI, but one has diabetes and one doesn't).
- One solution: Make the decision tree "less complex" by limiting the maximum depth.
Since sklearn.tree's plot_tree can't visualize extremely large decision trees, let's create and visualize some smaller decision trees.
trees = {}
for d in [2, 4, 8]:
trees[d] = DecisionTreeClassifier(max_depth=d, random_state=1)
trees[d].fit(X_train, y_train)
plt.figure(figsize=(15, 5), dpi=100)
plot_tree(trees[d], feature_names=X_train.columns, class_names=['no db', 'yes db'],
filled=True, rounded=True, impurity=False)
plt.show()
As tree depth increases, complexity increases, and our trees are more prone to overfitting. This means model bias decreases, but model variance increases.
Question: What is the "right" maximum depth to choose?
Hyperparameters for decision trees¶
max_depthis a hyperparameter forDecisionTreeClassifier.
- There are many more hyperparameters we can tweak; look at the documentation for examples.
min_samples_split: The minimum number of samples required to split an internal node.criterion: The function to measure the quality of a split ('gini'or'entropy').
- To ensure that our model generalizes well to unseen data, we need an efficient technique for trying different combinations of hyperparameters!
- We'll see that technique soon:
GridSearchCV.
Thinking about bias and variance¶
- Bigger
max_depth= less bias, more variance. - Bigger
min_samples_split= more bias, less variance. (Why?)
Question 🤔 (Answer at dsc80.com/q)
(Fa23 Final 10.1)
Suppose we fit decision trees of varying depths to predict 'y' using 'x1' and 'x2'. The entire training set is shown in the table below.
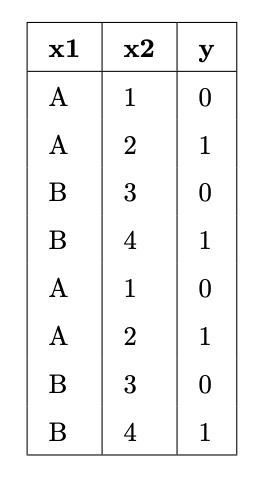
What is:
- The entropy of a node containing all the training points.
- The lowest possible entropy of a node in a fitted tree with depth 1 (two leaf nodes).
- The lowest possible entropy of a node in a fitted tree with depth 2 (four leaf nodes).
Grid search¶
Grid search¶
GridSearchCV takes in:
- an un-
fitinstance of an estimator, and - a dictionary of hyperparameter values to try,
and performs $k$-fold cross-validation to find the combination of hyperparameters with the best average validation performance.
from sklearn.model_selection import GridSearchCV
The following dictionary contains the values we're considering for each hyperparameter. (We're using GridSearchCV with 3 hyperparameters, but we could use it with even just a single hyperparameter.)
hyperparameters = {
'max_depth': [2, 3, 4, 5, 7, 10, 13, 15, 18, None],
'min_samples_split': [2, 5, 10, 20, 50, 100, 200],
'criterion': ['gini', 'entropy']
}
Note that there are 140 combinations of hyperparameters we need to try. We need to find the best combination of hyperparameters, not the best value for each hyperparameter individually.
len(hyperparameters['max_depth']) * \
len(hyperparameters['min_samples_split']) * \
len(hyperparameters['criterion'])
140
GridSearchCV needs to be instantiated and fit.
searcher = GridSearchCV(DecisionTreeClassifier(), hyperparameters, cv=5)
%%time
searcher.fit(X_train, y_train)
CPU times: user 853 ms, sys: 2.17 ms, total: 855 ms Wall time: 858 ms
GridSearchCV(cv=5, estimator=DecisionTreeClassifier(),
param_grid={'criterion': ['gini', 'entropy'],
'max_depth': [2, 3, 4, 5, 7, 10, 13, 15, 18, None],
'min_samples_split': [2, 5, 10, 20, 50, 100, 200]})In a Jupyter environment, please rerun this cell to show the HTML representation or trust the notebook. On GitHub, the HTML representation is unable to render, please try loading this page with nbviewer.org.
GridSearchCV(cv=5, estimator=DecisionTreeClassifier(),
param_grid={'criterion': ['gini', 'entropy'],
'max_depth': [2, 3, 4, 5, 7, 10, 13, 15, 18, None],
'min_samples_split': [2, 5, 10, 20, 50, 100, 200]})DecisionTreeClassifier(max_depth=4, min_samples_split=50)
DecisionTreeClassifier(max_depth=4, min_samples_split=50)
After being fit, the best_params_ attribute provides us with the best combination of hyperparameters to use.
searcher.best_params_
{'criterion': 'gini', 'max_depth': 4, 'min_samples_split': 50}
All of the intermediate results – validation accuracies for each fold, mean validation accuaries, etc. – are stored in the cv_results_ attribute:
searcher.cv_results_['mean_test_score'] # Array of length 140.
array([0.73, 0.73, 0.73, ..., 0.75, 0.74, 0.72])
# Rows correspond to folds, columns correspond to hyperparameter combinations.
pd.DataFrame(np.vstack([searcher.cv_results_[f'split{i}_test_score'] for i in range(5)]))
| 0 | 1 | 2 | 3 | ... | 136 | 137 | 138 | 139 | |
|---|---|---|---|---|---|---|---|---|---|
| 0 | 0.71 | 0.71 | 0.71 | 0.71 | ... | 0.67 | 0.70 | 0.71 | 0.73 |
| 1 | 0.77 | 0.77 | 0.77 | 0.77 | ... | 0.82 | 0.83 | 0.77 | 0.76 |
| 2 | 0.74 | 0.74 | 0.74 | 0.74 | ... | 0.68 | 0.72 | 0.74 | 0.73 |
| 3 | 0.70 | 0.70 | 0.70 | 0.70 | ... | 0.77 | 0.79 | 0.76 | 0.70 |
| 4 | 0.72 | 0.72 | 0.72 | 0.72 | ... | 0.70 | 0.71 | 0.72 | 0.70 |
5 rows × 140 columns
Note that the above DataFrame tells us that 5 * 140 = 700 models were trained in total!
Now that we've found the best combination of hyperparameters, we should fit a decision tree instance using those hyperparameters on our entire training set.
searcher.best_params_
{'criterion': 'gini', 'max_depth': 4, 'min_samples_split': 50}
final_tree = DecisionTreeClassifier(**searcher.best_params_)
final_tree
DecisionTreeClassifier(max_depth=4, min_samples_split=50)In a Jupyter environment, please rerun this cell to show the HTML representation or trust the notebook.
On GitHub, the HTML representation is unable to render, please try loading this page with nbviewer.org.
DecisionTreeClassifier(max_depth=4, min_samples_split=50)
final_tree.fit(X_train, y_train)
DecisionTreeClassifier(max_depth=4, min_samples_split=50)In a Jupyter environment, please rerun this cell to show the HTML representation or trust the notebook.
On GitHub, the HTML representation is unable to render, please try loading this page with nbviewer.org.
DecisionTreeClassifier(max_depth=4, min_samples_split=50)
# Training accuracy.
final_tree.score(X_train, y_train)
0.7881944444444444
# Testing accuracy.
# A bit lower than the `dt` tree we fit above!
final_tree.score(X_test, y_test)
0.765625
Remember, searcher itself is a model object (we had to fit it). After performing $k$-fold cross-validation, behind the scenes, searcher is trained on the entire training set using the optimal combination of hyperparameters.
In other words, searcher makes the same predictions that final_tree does!
searcher.score(X_train, y_train)
0.7881944444444444
searcher.score(X_test, y_test)
0.765625
Choosing possible hyperparameter values¶
A full grid search can take a long time.
- In our previous example, we tried 140 combinations of hyperparameters.
- Since we performed 5-fold cross-validation, we trained 700 decision trees under the hood.
Question: How do we pick the possible hyperparameter values to try?
Answer: Trial and error.
- If the "best" choice of a hyperparameter was at an extreme, try increasing the range.
- For instance, if you try
max_depths from 32 to 128, and 32 was the best, try includingmax_depthsunder 32.
Key takeaways¶
- Decision trees are trained by finding the best questions to ask using the features in the training data. A good question is one that isolates classes as much as possible.
- Decision trees have a tendency to overfit to training data. One way to mitigate this is by restricting the maximum depth of the tree.
- To efficiently find hyperparameters through cross-validation, use
GridSearchCV.- Specify which values to try for each hyperparameter, and
GridSearchCVwill try all unique combinations of hyperparameters and return the combination with the best average validation performance. GridSearchCVis not the only solution – seeRandomizedSearchCVif you're curious.
- Specify which values to try for each hyperparameter, and
Decision tree pros and cons¶
Pros:
- Fast to fit (usually log-linear time w.r.t. size of design matrix)
- Fast to predict (usually log time)
- Interpretable
- Robust to irrelevant features (think about why!)
- Linear transformations on features don't affect predictions (think about why!)
Cons:
- High variance: complete tree (no depth limit) will almost always overfit!
- Aren't the best at prediction in general.
Random Forests¶
Another idea:¶
Train a bunch of decision trees, then have them vote on a prediction!
- Problem: If you use the same training data, you will always get the same tree.
- Solution: Introduce randomness into training procedure to get different trees.
Idea 1: Bootstrap the training data¶
- We can bootstrap the training data $T$ times, then train one tree on each resample.
- Also known as bagging (Bootstrap AGgregating). In general, combining different predictors together is a useful technique called ensemble learning.
- For decision trees though, doesn't make trees different enough from each other (e.g. if you have one really strong predictor, it'll always be the first split).
Idea 2: Only use a subset of features¶
At each split, take a random subset of $ m $ features instead of choosing from all $ d $ of them.
Rule of thumb: $ m \approx \sqrt d $ seems to work well.
Key idea: For ensemble learning, you want the individual predictors to have low bias, high variance, and be uncorrelated with each other. That way, when you average them together, you have low bias AND low variance.
Random forest algorithm: Fit $ T $ trees by using bagging and a random subset of features at each split. Predict by taking a vote from the $ T $ trees.
Question 🤔 (Answer at dsc80.com/q)
Code: bvm
How will increasing $ m $ affect the bias / variance of each decision tree?
Example¶
# Let's use more features for prediction
from sklearn.model_selection import train_test_split
X_train, X_test, y_train, y_test = (
train_test_split(diabetes.drop(columns=['Outcome']), diabetes['Outcome'], random_state=1)
)
from sklearn.ensemble import RandomForestClassifier
clf = RandomForestClassifier()
clf.fit(X_train, y_train)
clf.score(X_train, y_train)
1.0
clf.score(X_test, y_test)
0.8125
Compared to our previous best decision tree with depth 4:
dt = DecisionTreeClassifier(max_depth=4, criterion='entropy')
dt.fit(X_train, y_train)
dt.score(X_train, y_train)
0.7829861111111112
dt.score(X_test, y_test)
0.734375
Example: Modeling using text features¶
Example: Fake news¶
We have a dataset containing news articles and labels for whether the article was deemed "fake" or "real". Credit to https://github.com/KaiDMML/FakeNewsNet.
news = pd.read_csv('data/fake_news_training.csv')
news
| baseurl | content | label | |
|---|---|---|---|
| 0 | twitter.com | \njavascript is not available.\n\nwe’ve detect... | real |
| 1 | whitehouse.gov | remarks by the president at campaign event -- ... | real |
| 2 | web.archive.org | the committee on energy and commerce\nbarton: ... | real |
| ... | ... | ... | ... |
| 658 | politico.com | full text: jeff flake on trump speech transcri... | fake |
| 659 | pol.moveon.org | moveon.org political action: 10 things to know... | real |
| 660 | uspostman.com | uspostman.com is for sale\nyes, you can transf... | fake |
661 rows × 3 columns
Goal: Use an article's content to predict its label.
news['label'].value_counts(normalize=True)
label real 0.55 fake 0.45 Name: proportion, dtype: float64
Question: What is the worst possible accuracy we should expect from a classifier, given the above distribution?
Aside: CountVectorizer¶
Entries in the 'content' column are not currently quantitative! We can use the bag of words encoding to create quantitative features out of each 'content'.
Instead of performing a bag of words encoding manually as we did before, we can rely on sklearn's CountVectorizer. (There is also a TfidfVectorizer.)
from sklearn.feature_extraction.text import CountVectorizer
example_corp = ['hey hey hey my name is billy',
'hey billy how is your dog billy']
count_vec = CountVectorizer()
count_vec.fit(example_corp)
CountVectorizer()In a Jupyter environment, please rerun this cell to show the HTML representation or trust the notebook.
On GitHub, the HTML representation is unable to render, please try loading this page with nbviewer.org.
CountVectorizer()
count_vec learned a vocabulary from the corpus we fit it on.
count_vec.vocabulary_
{'hey': 2,
'my': 5,
'name': 6,
'is': 4,
'billy': 0,
'how': 3,
'your': 7,
'dog': 1}
count_vec.transform(example_corp).toarray()
array([[1, 0, 3, 0, 1, 1, 1, 0],
[2, 1, 1, 1, 1, 0, 0, 1]])
Note that the values in count_vec.vocabulary_ correspond to the positions of the columns in count_vec.transform(example_corp).toarray(), i.e. 'billy' is the first column and 'your' is the last column.
example_corp
['hey hey hey my name is billy', 'hey billy how is your dog billy']
pd.DataFrame(count_vec.transform(example_corp).toarray(),
columns=pd.Series(count_vec.vocabulary_).sort_values().index)
| billy | dog | hey | how | is | my | name | your | |
|---|---|---|---|---|---|---|---|---|
| 0 | 1 | 0 | 3 | 0 | 1 | 1 | 1 | 0 |
| 1 | 2 | 1 | 1 | 1 | 1 | 0 | 0 | 1 |
Creating an initial Pipeline¶
Let's build a Pipeline that takes in summaries and overall ratings and:
Uses
CountVectorizerto quantitatively encode summaries.Fits a
RandomForestClassifierto the data.
But first, a train-test split (like always).
from sklearn.model_selection import train_test_split
from sklearn.pipeline import Pipeline
from sklearn.ensemble import RandomForestClassifier
X = news['content']
y = news['label']
X_train, X_test, y_train, y_test = train_test_split(X, y)
To start, we'll create a random forest with 100 trees (n_estimators) each of which has a maximum depth of 3 (max_depth).
pl = Pipeline([
('cv', CountVectorizer()),
('clf', RandomForestClassifier(
max_depth=3,
n_estimators=100, # Uses 100 separate decision trees!
random_state=42,
))
])
pl.fit(X_train, y_train)
Pipeline(steps=[('cv', CountVectorizer()),
('clf', RandomForestClassifier(max_depth=3, random_state=42))])In a Jupyter environment, please rerun this cell to show the HTML representation or trust the notebook. On GitHub, the HTML representation is unable to render, please try loading this page with nbviewer.org.
Pipeline(steps=[('cv', CountVectorizer()),
('clf', RandomForestClassifier(max_depth=3, random_state=42))])CountVectorizer()
RandomForestClassifier(max_depth=3, random_state=42)
# Training accuracy.
pl.score(X_train, y_train)
0.7616161616161616
# Testing accuracy.
pl.score(X_test, y_test)
0.6807228915662651
The accuracy of our random forest is just under 73%, on the test set. How much better does it do compared to a classifier that predicts "real" every time?
y_train.value_counts(normalize=True)
label real 0.54 fake 0.46 Name: proportion, dtype: float64
# Distribution of predicted ys in the training set:
# stops scientific notation for pandas
pd.set_option('display.float_format', '{:.3f}'.format)
pd.Series(pl.predict(X_train)).value_counts(normalize=True)
fake 0.681 real 0.319 Name: proportion, dtype: float64
len(pl.named_steps['cv'].vocabulary_) # Lots of features!
23079
Choosing tree depth via GridSearchCV¶
We arbitrarily chose max_depth=3 before, but it seems like that isn't working well. Let's perform a grid search to find the max_depth with the best generalization performance.
# Note that we've used the key clf__max_depth, not max_depth
# because max_depth is a hyperparameter of clf, not of pl.
hyperparameters = {
'clf__max_depth': np.arange(2, 200, 20)
}
Note that while pl has already been fit, we can still give it to GridSearchCV, which will repeatedly re-fit it during cross-validation.
%%time
# Takes a few seconds to run – how many trees are being trained?
from sklearn.model_selection import GridSearchCV
grids = GridSearchCV(
pl,
n_jobs=-1, # Use multiple processors to parallelize
param_grid=hyperparameters,
return_train_score=True
)
grids.fit(X_train, y_train)
CPU times: user 1.18 s, sys: 255 ms, total: 1.44 s Wall time: 6.4 s
GridSearchCV(estimator=Pipeline(steps=[('cv', CountVectorizer()),
('clf',
RandomForestClassifier(max_depth=3,
random_state=42))]),
n_jobs=-1,
param_grid={'clf__max_depth': array([ 2, 22, 42, 62, 82, 102, 122, 142, 162, 182])},
return_train_score=True)In a Jupyter environment, please rerun this cell to show the HTML representation or trust the notebook. On GitHub, the HTML representation is unable to render, please try loading this page with nbviewer.org.
GridSearchCV(estimator=Pipeline(steps=[('cv', CountVectorizer()),
('clf',
RandomForestClassifier(max_depth=3,
random_state=42))]),
n_jobs=-1,
param_grid={'clf__max_depth': array([ 2, 22, 42, 62, 82, 102, 122, 142, 162, 182])},
return_train_score=True)Pipeline(steps=[('cv', CountVectorizer()),
('clf',
RandomForestClassifier(max_depth=np.int64(42),
random_state=42))])CountVectorizer()
RandomForestClassifier(max_depth=np.int64(42), random_state=42)
grids.best_params_
{'clf__max_depth': np.int64(42)}
Recall, fit GridSearchCV objects are estimators on their own as well. This means we can compute the training and testing accuracies of the "best" random forest directly:
# Training accuracy.
grids.score(X_train, y_train)
0.9959595959595959
# Testing accuracy.
grids.score(X_test, y_test)
0.8373493975903614
~10% better test set error!
Training and validation accuracy vs. depth¶
Below, we plot how training and validation accuracy varied with tree depth. Note that the $y$-axis here is accuracy, and that larger accuracies are better (unlike with RMSE, where smaller was better).
index = grids.param_grid['clf__max_depth']
train = grids.cv_results_['mean_train_score']
valid = grids.cv_results_['mean_test_score']
pd.DataFrame({'train': train, 'valid': valid}, index=index).plot().update_layout(
xaxis_title='max_depth', yaxis_title='Accuracy'
)
Question 🤔 (Answer at dsc80.com/q)
Code: fa23102
(Fa23 Final Q10.2)
Suppose we write the following code:
hyperparameters = {
'n_estimators': [10, 100, 1000], # number of trees per forest
'max_depth': [None, 100, 10] # max depth of each tree
}
grids = GridSearchCV(
RandomForestClassifier(), param_grid=hyperparameters,
cv=3, # 3-fold cross-validation
)
grids.fit(X_train, y_train)
Answer the following questions with a single number.
- How many random forests are fit in total?
- How many decision trees are fit in total?
- How many times in total is the first point in X_train used to train a decision tree?
Classifier Evaluation¶
Accuracy isn't everything!¶
$$ \text{accuracy} = \frac{\text{\# data points classified correctly}}{\text{\# data points}} $$
Accuracy is defined as the proportion of predictions that are correct.
It weighs all correct predictions the same, and weighs all incorrect predictions the same.
But some incorrect predictions may be worse than others!
- Example: Suppose you take a COVID test 🦠. Which is worse:
- The test saying you have COVID, when you really don't, or
- The test saying you don't have COVID, when you really do?
- Example: Suppose you take a COVID test 🦠. Which is worse:
Repeat the previous paragraph many, many times.
One night, the shepherd boy sees a real wolf approaching the flock and calls out, "Wolf!" The villagers refuse to be fooled again and stay in their houses. The hungry wolf turns the flock into lamb chops. The town goes hungry. Panic ensues.
The wolf classifier¶
- Predictor: Shepherd boy.
- Positive prediction: "There is a wolf."
- Negative prediction: "There is no wolf."
Some questions to think about:
- What is an example of an incorrect, positive prediction?
- Was there a correct, negative prediction?
- There are four possibilities. What are the consequences of each?
- (predict yes, predict no) x (actually yes, actually no).
The wolf classifier¶
Below, we present a confusion matrix, which summarizes the four possible outcomes of the wolf classifier.
Outcomes in binary classification¶
When performing binary classification, there are four possible outcomes.
(Note: A "positive prediction" is a prediction of 1, and a "negative prediction" is a prediction of 0.)
| Outcome of Prediction | Definition | True Class |
|---|---|---|
| True positive (TP) ✅ | The predictor correctly predicts the positive class. | P |
| False negative (FN) ❌ | The predictor incorrectly predicts the negative class. | P |
| True negative (TN) ✅ | The predictor correctly predicts the negative class. | N |
| False positive (FP) ❌ | The predictor incorrectly predicts the positive class. | N |
| Predicted Negative | Predicted Positive | |
|---|---|---|
| Actually Negative | TN ✅ | FP ❌ |
| Actually Positive | FN ❌ | TP ✅ |
sklearn's confusion matrices are (but differently than in the wolf example).Note that in the four acronyms – TP, FN, TN, FP – the first letter is whether the prediction is correct, and the second letter is what the prediction is.
Example: COVID testing 🦠¶
UCSD Health administers hundreds of COVID tests a day. The tests are not fully accurate.
Each test comes back either
- positive, indicating that the individual has COVID, or
- negative, indicating that the individual does not have COVID.
Question: What is a TP in this scenario? FP? TN? FN?
TP: The test predicted that the individual has COVID, and they do ✅.
FP: The test predicted that the individual has COVID, but they don't ❌.
TN: The test predicted that the individual doesn't have COVID, and they don't ✅.
FN: The test predicted that the individual doesn't have COVID, but they do ❌.
Accuracy of COVID tests¶
The results of 100 UCSD Health COVID tests are given below.
| Predicted Negative | Predicted Positive | |
|---|---|---|
| Actually Negative | TN = 90 ✅ | FP = 1 ❌ |
| Actually Positive | FN = 8 ❌ | TP = 1 ✅ |
🤔 Question: What is the accuracy of the test?
🙋 Answer: $$\text{accuracy} = \frac{TP + TN}{TP + FP + FN + TN} = \frac{1 + 90}{100} = 0.91$$
Followup: At first, the test seems good. But, suppose we build a classifier that predicts that nobody has COVID. What would its accuracy be?
Answer to followup: Also 0.91! There is severe class imbalance in the dataset, meaning that most of the data points are in the same class (no COVID). Accuracy doesn't tell the full story.
Recall¶
| Predicted Negative | Predicted Positive | |
|---|---|---|
| Actually Negative | TN = 90 ✅ | FP = 1 ❌ |
| Actually Positive | FN = 8 ❌ | TP = 1 ✅ |
🤔 Question: What proportion of individuals who actually have COVID did the test identify?
🙋 Answer: $\frac{1}{1 + 8} = \frac{1}{9} \approx 0.11$
More generally, the recall of a binary classifier is the proportion of actually positive instances that are correctly classified. We'd like this number to be as close to 1 (100%) as possible.
$$\text{recall} = \frac{TP}{\text{\# actually positive}} = \frac{TP}{TP + FN}$$
To compute recall, look at the bottom (positive) row of the above confusion matrix.
Recall isn't everything, either!¶
$$\text{recall} = \frac{TP}{TP + FN}$$
🤔 Question: Can you design a "COVID test" with perfect recall?
🙋 Answer: Yes – just predict that everyone has COVID!
| Predicted Negative | Predicted Positive | |
|---|---|---|
| Actually Negative | TN = 0 ✅ | FP = 91 ❌ |
| Actually Positive | FN = 0 ❌ | TP = 9 ✅ |
$$\text{recall} = \frac{TP}{TP + FN} = \frac{9}{9 + 0} = 1$$
Like accuracy, recall on its own is not a perfect metric. Even though the classifier we just created has perfect recall, it has 91 false positives!
Precision¶
| Predicted Negative | Predicted Positive | |
|---|---|---|
| Actually Negative | TN = 0 ✅ | FP = 91 ❌ |
| Actually Positive | FN = 0 ❌ | TP = 9 ✅ |
The precision of a binary classifier is the proportion of predicted positive instances that are correctly classified. We'd like this number to be as close to 1 (100%) as possible.
$$\text{precision} = \frac{TP}{\text{\# predicted positive}} = \frac{TP}{TP + FP}$$
To compute precision, look at the right (positive) column of the above confusion matrix.
Tip: A good way to remember the difference between precision and recall is that in the denominator for 🅿️recision, both terms have 🅿️ in them (TP and FP).
Note that the "everyone-has-COVID" classifier has perfect recall, but a precision of $\frac{9}{9 + 91} = 0.09$, which is quite low.
🚨 Key idea: There is a "tradeoff" between precision and recall. Ideally, you want both to be high. For a particular prediction task, one may be important than the other.
Precision and recall¶
$$\text{precision} = \frac{TP}{TP + FP} \: \: \: \: \: \: \: \: \text{recall} = \frac{TP}{TP + FN}$$
Question 🤔 (Answer at dsc80.com/q)
Code: pvsr
🤔 When might high precision be more important than high recall?
🤔 When might high recall be more important than high precision?
Question 🤔 (Answer at dsc80.com/q)
Code: apr
Consider the confusion matrix shown below.
| Predicted Negative | Predicted Positive | |
|---|---|---|
| Actually Negative | TN = 22 ✅ | FP = 2 ❌ |
| Actually Positive | FN = 23 ❌ | TP = 18 ✅ |
What is the accuracy of the above classifier? The precision? The recall?
Summary, next time¶
Summary¶
- Decision trees, while interpretable, are prone to having high variance. There are several ways to control the variance of a decision tree:
- Limit
max_depthor increasemin_samples_split. - Create a random forest, which is an ensemble of multiple decision trees, each fit to a different random resample of the training data, using a random sample of features.
- Limit
- In order to tune model hyperparameters – that is, to find the hyperparameters that likely maximize performance on unseen data – use
GridSearchCV. - Accuracy alone is not always a meaningful representation of a classifier's quality, particularly when the classes are imbalanced.
- Precision and recall are classifier evaluation metrics that consider the types of errors being made.
- There is a "tradeoff" between precision and recall. One may be more important than the other, depending on the task.