from dsc80_utils import *
# The dataset is built into plotly (and seaborn)!
# We shuffle here so that the head of the DataFrame contains rows where smoker is Yes and smoker is No,
# purely for illustration purposes (it doesn't change any of the math).
np.random.seed(1)
tips = px.data.tips().sample(frac=1).reset_index(drop=True)
Announcements 📣¶
- Lab 7 is due tomorrow.
- The Final Project is out!
- It will be worth two projects (because it used to be two separate projects).
- It will have two short checkpoints due this Friday and next Friday.
- You can request an extension on the checkpoints.
- You cannot request an extension on final submission deadline on Wednesday, June 12 (the Wed of finals week).
Agenda 📆¶
- Review: Predicting tips.
- $R^2$.
- Feature engineering.
- Example: Predicting tips.
- One hot encoding.
- Example: Predicting ratings ⭐️.
- Dropping features.
- Ordinal encoding.
- Example: Horsepower 🚗.
- Quantitative scaling.
- Example: Predicting tips.
- Feature engineering in
sklearn.- Transformer classes.
- Creating
Pipelines.
Review: Predicting tips 🧑🍳¶
tips
| total_bill | tip | sex | smoker | day | time | size | |
|---|---|---|---|---|---|---|---|
| 0 | 3.07 | 1.00 | Female | Yes | Sat | Dinner | 1 |
| 1 | 18.78 | 3.00 | Female | No | Thur | Dinner | 2 |
| 2 | 26.59 | 3.41 | Male | Yes | Sat | Dinner | 3 |
| ... | ... | ... | ... | ... | ... | ... | ... |
| 241 | 17.47 | 3.50 | Female | No | Thur | Lunch | 2 |
| 242 | 10.07 | 1.25 | Male | No | Sat | Dinner | 2 |
| 243 | 16.93 | 3.07 | Female | No | Sat | Dinner | 3 |
244 rows × 7 columns
Linear models¶
Last time, we fit three linear models to predict restaurant tips:
- Constant model: $\text{predicted tip} = h$.
- Simple linear regression: $\text{predicted tip} = w_0 + w_1 \cdot \text{total bill}$.
- Multiple linear regression: $\text{predicted tip} = w_0 + w_1 \cdot \text{total bill} + w_2 \cdot \text{table size}$.
In the constant model case, we know that the optimal model parameter, when using squared loss, is $h^* = \text{mean tip}$.
mean_tip = tips['tip'].mean()
In the other two cases, we used the LinearRegression class from sklearn to help us find optimal model parameters.
from sklearn.linear_model import LinearRegression
model = LinearRegression()
model.fit(X=tips[['total_bill']], y=tips['tip'])
model_two = LinearRegression()
model_two.fit(X=tips[['total_bill', 'size']], y=tips['tip'])
LinearRegression()
Root mean squared error¶
To compare the performance of different models, we used the root mean squared error (RMSE).
$$\text{RMSE} = \sqrt{\frac{1}{n} \sum_{i = 1}^n \big( y_i - H(x_i) \big)^2}$$def rmse(actual, pred):
return np.sqrt(np.mean((actual - pred) ** 2))
rmse_dict = {}
rmse_dict['constant tip amount'] = rmse(tips['tip'], mean_tip)
all_preds = model.predict(tips[['total_bill']])
rmse_dict['one feature: total bill'] = rmse(tips['tip'], all_preds)
rmse_dict['two features'] = rmse(
tips['tip'], model_two.predict(tips[['total_bill', 'size']])
)
pd.DataFrame({'rmse': rmse_dict.values()}, index=rmse_dict.keys())
| rmse | |
|---|---|
| constant tip amount | 1.38 |
| one feature: total bill | 1.02 |
| two features | 1.01 |
The .score method of a LinearRegression object¶
Model objects in sklearn that have already been fit have a score method.
model_two.score(tips[['total_bill', 'size']], tips['tip'])
0.46786930879612565
That doesn't look like the RMSE... what is it? 🤔
Aside: $R^2$¶
- $R^2$, or the coefficient of determination, is a measure of the quality of a linear fit.
- There are a few equivalent ways of computing it, assuming your model is linear and has an intercept term:
- Interpretation: $R^2$ is the proportion of variance in $y$ that the linear model explains.
- In the simple linear regression case, it is the square of the correlation coefficient, $r$.
- Key idea: $R^2$ ranges from 0 to 1. The closer it is to 1, the better the linear fit is.
- $R^2$ has no units of measurement, unlike RMSE.
Calculating $R^2$¶
Let's calculate the $R^2$ for model_two's predictions in three different ways.
pred = tips.assign(predicted=model_two.predict(tips[['total_bill', 'size']]))
pred
| total_bill | tip | sex | smoker | day | time | size | predicted | |
|---|---|---|---|---|---|---|---|---|
| 0 | 3.07 | 1.00 | Female | Yes | Sat | Dinner | 1 | 1.15 |
| 1 | 18.78 | 3.00 | Female | No | Thur | Dinner | 2 | 2.80 |
| 2 | 26.59 | 3.41 | Male | Yes | Sat | Dinner | 3 | 3.71 |
| ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 241 | 17.47 | 3.50 | Female | No | Thur | Lunch | 2 | 2.67 |
| 242 | 10.07 | 1.25 | Male | No | Sat | Dinner | 2 | 1.99 |
| 243 | 16.93 | 3.07 | Female | No | Sat | Dinner | 3 | 2.82 |
244 rows × 8 columns
Method 1: $R^2 = \frac{\text{var}(\text{predicted $y$ values})}{\text{var}(\text{actual $y$ values})}$
np.var(pred['predicted']) / np.var(pred['tip'])
0.4678693087961255
Method 2: $R^2 = \left[ \text{correlation}(\text{predicted $y$ values}, \text{actual $y$ values}) \right]^2$
Note: By correlation here, we are referring to $r$, the same correlation coefficient you saw in DSC 10.
pred.corr().loc['predicted', 'tip'] ** 2
0.46786930879612554
Method 3: LinearRegression.score
model_two.score(tips[['total_bill', 'size']], tips['tip'])
0.46786930879612565
All three methods provide the same result!
Relationship between $R^2$ and RMSE¶
For linear models with an intercept term,
$$R^2 = 1 - \frac{\text{RMSE}^2}{\text{var}(\text{actual $y$ values})}$$1 - rmse(pred['tip'], pred['predicted']) ** 2 / np.var(pred['tip'])
0.4678693087961261
What's next?¶
tips.head()
| total_bill | tip | sex | smoker | day | time | size | |
|---|---|---|---|---|---|---|---|
| 0 | 3.07 | 1.00 | Female | Yes | Sat | Dinner | 1 |
| 1 | 18.78 | 3.00 | Female | No | Thur | Dinner | 2 |
| 2 | 26.59 | 3.41 | Male | Yes | Sat | Dinner | 3 |
| 3 | 14.26 | 2.50 | Male | No | Thur | Lunch | 2 |
| 4 | 21.16 | 3.00 | Male | No | Thur | Lunch | 2 |
So far, in our journey to predict
'tip', we've only used the existing numerical features in our dataset,'total_bill'and'size'.There's a lot of information in tips that we didn't use –
'sex','smoker','day', and'time', for example. We can't use these features in their current form, because they're non-numeric.How do we use categorical features in a regression model?
Feature engineering ⚙️¶
The goal of feature engineering¶
- Feature engineering is the act of finding transformations that transform data into effective quantitative variables.
- A feature function $\phi$ (phi, pronounced "fea") is a mapping from raw data to $d$-dimensional space, i.e. $\phi: \text{raw data} \rightarrow \mathbb{R}^d$.
- If two observations $x_i$ and $x_j$ are "similar" in the raw data space, then $\phi(x_i)$ and $\phi(x_j)$ should also be "similar."
- A "good" choice of features depends on many factors:
- The kind of data, i.e. quantitative, ordinal, or nominal.
- The relationship(s) being modeled.
- The model type, e.g. linear models, decision tree models, neural networks.
- To introduce different feature functions, we'll look at several different example datasets.
One hot encoding¶
- One hot encoding is a transformation that turns a categorical feature into several binary features.
- Suppose a column has $N$ unique values, $A_1$, $A_2$, ..., $A_N$. For each unique value $A_i$, we define the following feature function:
- Note that 1 means "yes" and 0 means "no".
- One hot encoding is also called "dummy encoding", and $\phi(x)$ may also be referred to as an "indicator variable".
Example: One hot encoding 'smoker'¶
For each unique value of 'smoker' in our dataset, we must create a column for just that 'smoker'. (Remember, 'smoker' is 'Yes' when the table was in the smoking section of the restaurant and 'No' otherwise.)
tips.head()
| total_bill | tip | sex | smoker | day | time | size | |
|---|---|---|---|---|---|---|---|
| 0 | 3.07 | 1.00 | Female | Yes | Sat | Dinner | 1 |
| 1 | 18.78 | 3.00 | Female | No | Thur | Dinner | 2 |
| 2 | 26.59 | 3.41 | Male | Yes | Sat | Dinner | 3 |
| 3 | 14.26 | 2.50 | Male | No | Thur | Lunch | 2 |
| 4 | 21.16 | 3.00 | Male | No | Thur | Lunch | 2 |
tips['smoker'].value_counts()
No 151 Yes 93 Name: smoker, dtype: int64
(tips['smoker'] == 'Yes').astype(int).head()
0 1 1 0 2 1 3 0 4 0 Name: smoker, dtype: int64
for val in tips['smoker'].unique():
tips[f'smoker == {val}'] = (tips['smoker'] == val).astype(int)
tips.head()
| total_bill | tip | sex | smoker | ... | time | size | smoker == Yes | smoker == No | |
|---|---|---|---|---|---|---|---|---|---|
| 0 | 3.07 | 1.00 | Female | Yes | ... | Dinner | 1 | 1 | 0 |
| 1 | 18.78 | 3.00 | Female | No | ... | Dinner | 2 | 0 | 1 |
| 2 | 26.59 | 3.41 | Male | Yes | ... | Dinner | 3 | 1 | 0 |
| 3 | 14.26 | 2.50 | Male | No | ... | Lunch | 2 | 0 | 1 |
| 4 | 21.16 | 3.00 | Male | No | ... | Lunch | 2 | 0 | 1 |
5 rows × 9 columns
Model #4: Multiple linear regression using total bill, table size, and smoker status¶
Now that we've converted 'smoker' to a numerical variable, we can use it as input in a regression model. Here's the model we'll try to fit:
Subtlety: There's no need to use both 'smoker == No' and 'smoker == Yes'. If we know the value of one, we already know the value of the other. We can use either one.
model_three = LinearRegression()
model_three.fit(tips[['total_bill', 'size', 'smoker == Yes']], tips['tip'])
LinearRegression()
The following cell gives us our $w^*$s:
model_three.intercept_, model_three.coef_
(0.7090155167346053, array([ 0.09, 0.18, -0.08]))
Thus, our trained linear model to predict tips given total bills, table sizes, and smoker status (yes or no) is:
$$\text{predicted tip} = 0.709 + 0.094 \cdot \text{total bill} + 0.180 \cdot \text{table size} - 0.083 \cdot \text{smoker == Yes}$$Visualizing Model #4¶
Our new fit model is:
$$\text{predicted tip} = 0.709 + 0.094 \cdot \text{total bill} + 0.180 \cdot \text{table size} - 0.083 \cdot \text{smoker == Yes}$$To visualize our data and linear model, we'd need 4 dimensions:
- One for total bill
- One for table size
- One for
'smoker == Yes'. - One for tip.
Humans can't visualize in 4D, but there may be a solution. We know that 'smoker == Yes' only has two possible values, 1 or 0, so let's look at those cases separately.
Case 1: 'smoker == Yes' is 1, meaning that the table was in the smoking section.
Case 2: 'smoker == Yes' is 0, meaning that the table was not in the smoking section.
Key idea: These are two parallel planes in 3D, with different $z$-intercepts!
Note that the two planes are very close to one another – you'll have to zoom in to see the difference.
# pio.renderers.default = 'plotly_mimetype+notebook' # If it doesn't render, try uncommenting this.
XX, YY = np.mgrid[0:50:2, 0:8:1]
Z_0 = model_three.intercept_ + model_three.coef_[0] * XX + model_three.coef_[1] * YY + model_three.coef_[2] * 0
Z_1 = model_three.intercept_ + model_three.coef_[0] * XX + model_three.coef_[1] * YY + model_three.coef_[2] * 1
plane_0 = go.Surface(x=XX, y=YY, z=Z_0, colorscale='Greens')
plane_1 = go.Surface(x=XX, y=YY, z=Z_1, colorscale='Purples')
fig = go.Figure(data=[plane_0, plane_1])
tips_0 = tips[tips['smoker'] == 'No']
tips_1 = tips[tips['smoker'] == 'Yes']
fig.add_trace(go.Scatter3d(x=tips_0['total_bill'],
y=tips_0['size'],
z=tips_0['tip'], mode='markers', marker = {'color': 'green'}))
fig.add_trace(go.Scatter3d(x=tips_1['total_bill'],
y=tips_1['size'],
z=tips_1['tip'], mode='markers', marker = {'color': 'purple'}))
fig.update_layout(scene = dict(
xaxis_title='Total Bill',
yaxis_title='Table Size',
zaxis_title='Tip'),
title='Tip vs. Total Bill and Table Size (Green = Non-Smoking Section, Purple = Smoking Section)',
width=1000, height=800,
showlegend=False)
If we want to visualize in 2D, we need to pick a single feature to place on the $x$-axis.
fig = go.Figure()
fig.add_trace(go.Scatter(x=tips['total_bill'], y=tips['tip'],
mode='markers', name='Original Data'))
fig.add_trace(go.Scatter(x=tips['total_bill'], y=model_three.predict(tips[['total_bill', 'size', 'smoker == Yes']]),
mode='markers', name='Predicted Tips using Total Bill, <br>Table Size, and Smoker Status'))
fig.update_layout(showlegend=True, title='Tip vs. Total Bill',
xaxis_title='Total Bill', yaxis_title='Tip')
Despite being a linear model, why doesn't this model look like a straight line?
Comparing Model #4 to earlier models¶
rmse_dict['three features'] = rmse(tips['tip'],
model_three.predict(tips[['total_bill', 'size', 'smoker == Yes']]))
rmse_dict
{'constant tip amount': 1.3807999538298956,
'one feature: total bill': 1.0178504025697377,
'two features': 1.0072561271146618,
'three features': 1.0064899786822126}
Adding 'smoker == Yes' decreased the training RMSE of our model, but barely.
Reflection¶
tips.head()
| total_bill | tip | sex | smoker | ... | time | size | smoker == Yes | smoker == No | |
|---|---|---|---|---|---|---|---|---|---|
| 0 | 3.07 | 1.00 | Female | Yes | ... | Dinner | 1 | 1 | 0 |
| 1 | 18.78 | 3.00 | Female | No | ... | Dinner | 2 | 0 | 1 |
| 2 | 26.59 | 3.41 | Male | Yes | ... | Dinner | 3 | 1 | 0 |
| 3 | 14.26 | 2.50 | Male | No | ... | Lunch | 2 | 0 | 1 |
| 4 | 21.16 | 3.00 | Male | No | ... | Lunch | 2 | 0 | 1 |
5 rows × 9 columns
We've one hot encoded
'smoker', but it required afor-loop.Is there an easy way to one hot encode all four categorical columns –
'sex','smoker','day', and'time'– all at once, without using afor-loop?Yes, using
sklearn.preprocessing'sOneHotEncoder. More on this soon!
Question 🤔 (Answer at q.dsc80.com)
(Fa23 Final Q9.1)
Every week, Lauren goes to her local grocery store and buys a varying amount of vegetable but always buys exactly one pound of meat (either beef, fish, or chicken). We use a linear regression model to predict her total grocery bill. We’ve collected a dataset containing the pounds of vegetables bought, the type of meat bought, and the total bill. Below we display the first few rows of the dataset and two plots generated using the entire training set.
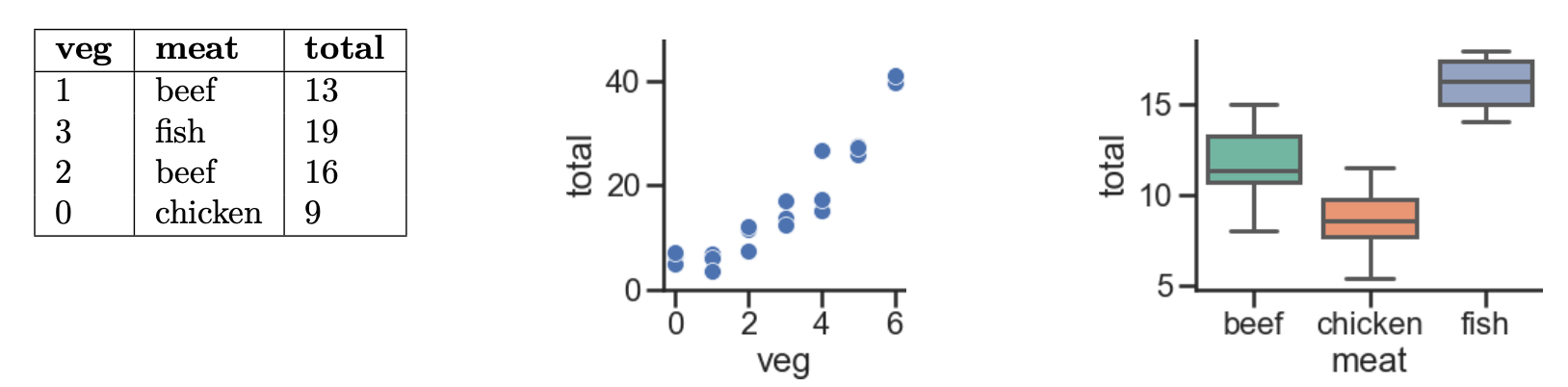
Suppose we fit the following linear regression models to predict
total. Based on the data and visualizations shown above, determine
whether the fitted model weights are positive (+), negative (-), or exactly
0. The notation meat=beef refers to the one-hot encoded meat
column with value 1 if the original value in the meat column was
beef and 0 otherwise. Likewise, meat=chicken and
meat=fish are the one-hot encoded meat columns for
chicken and fish, respectively.
- $H(x) = w_0 $
- $H(x) = w_0 + w_1 \cdot \text{veg} $
- $H(x) = w_0 + w_1 \cdot (\text{meat=chicken}) $
- $H(x) = w_0 + w_1 \cdot (\text{meat=beef}) + w_2 \cdot (\text{meat=chicken}) $
- $H(x) = w_0 + w_1 \cdot (\text{meat=beef}) + w_2 \cdot (\text{meat=chicken}) + w_3 \cdot (\text{meat=fish}) $
Example: Predicting ratings ⭐️¶
Example: Predicting ratings ⭐️¶
| UID | AGE | STATE | HAS_BOUGHT | REVIEW | \ | |
|---|---|---|---|---|---|---|
| 74 | 32 | NY | True | "Meh." | | | ✩✩ |
| 42 | 50 | WA | True | "Worked out of the box..." | | | ✩✩✩✩ |
| 57 | 16 | CA | NULL | "Sick af..." | | | ✩ |
| ... | ... | ... | ... | ... | | | ... |
| (int) | (int) | (str) | (bool) | (str) | | | (str) |
- We want to build a classifier that predicts
'RATING'using the above features.
- Why can't we build a model right away? What must we do so that we can build a model?
- Some issues: missing values, emojis and strings instead of numbers, irrelevant columns.
Uninformative features¶
'UID'was likely used to join the user information (e.g.,'AGE'and'STATE') with somereviewsdataset.
- Even though
'UID's are stored as numbers, the numerical value of a user's'UID'won't help us predict their'RATING'.
- If we include the
'UID'feature, our model will find whatever patterns it can between'UID's and'RATING's in the training (observed data).- This will lead to a lower training RMSE.
- However, since there is truly no relationship between
'UID'and'RATING', this will lead to worse model performance on unseen data (bad).
Dropping features¶
There are certain scenarios where manually dropping features might be helpful:
- When the features do not contain information associated with the prediction task.
- When the feature is not available at prediction time.
- The goal of building a model to predict
'RATING's is so that we can predict'RATING's for users who haven't actually made a'RATING'yet.
- As such, our model should only depend on features that we would know before the user makes their
'RATING'.
- For instance, if a user only enters a
'REVIEW'after entering a'RATING', we shouldn't use their'REVIEW'to predict their'RATING'.
Encoding ordinal features¶
| UID | AGE | STATE | HAS_BOUGHT | REVIEW | \ | |
|---|---|---|---|---|---|---|
| 74 | 32 | NY | True | "Meh." | | | ✩✩ |
| 42 | 50 | WA | True | "Worked out of the box..." | | | ✩✩✩✩ |
| 57 | 16 | CA | NULL | "Sick af..." | | | ✩ |
| ... | ... | ... | ... | ... | | | ... |
| (int) | (int) | (str) | (bool) | (str) | | | (str) |
How do we encode the 'RATING' column, an ordinal variable, as a quantitative variable?
- Transformation: Replace "number of ✩" with "number".
- This is an ordinal encoding, a transformation that maps ordinal values to the positive integers in a way that preserves order.
- Example: (freshman, sophomore, junior, senior) -> (0, 1, 2, 3).
- Important: This transformation preserves "distances" between ratings.
ordinal_enc = {
'✩': 1,
'✩✩': 2,
'✩✩✩': 3,
'✩✩✩✩': 4,
'✩✩✩✩✩': 5,
}
ordinal_enc
{'✩': 1, '✩✩': 2, '✩✩✩': 3, '✩✩✩✩': 4, '✩✩✩✩✩': 5}
ratings = pd.DataFrame().assign(RATING=['✩', '✩✩', '✩✩✩', '✩✩', '✩✩✩', '✩', '✩✩✩', '✩✩✩✩', '✩✩✩✩✩'])
ratings
| RATING | |
|---|---|
| 0 | ✩ |
| 1 | ✩✩ |
| 2 | ✩✩✩ |
| ... | ... |
| 6 | ✩✩✩ |
| 7 | ✩✩✩✩ |
| 8 | ✩✩✩✩✩ |
9 rows × 1 columns
ratings.replace(ordinal_enc)
| RATING | |
|---|---|
| 0 | 1 |
| 1 | 2 |
| 2 | 3 |
| ... | ... |
| 6 | 3 |
| 7 | 4 |
| 8 | 5 |
9 rows × 1 columns
Encoding nominal features¶
| UID | AGE | STATE | HAS_BOUGHT | REVIEW | \ | |
|---|---|---|---|---|---|---|
| 74 | 32 | NY | True | "Meh." | | | ✩✩ |
| 42 | 50 | WA | True | "Worked out of the box..." | | | ✩✩✩✩ |
| 57 | 16 | CA | NULL | "Sick af..." | | | ✩ |
| ... | ... | ... | ... | ... | | | ... |
| (int) | (int) | (str) | (bool) | (str) | | | (str) |
How do we encode the 'STATE' column, a nominal variable, as a quantitative variable? In other words, how do we turn 'STATE's into meaningful numbers?
- Question: Why can't we use an ordinal encoding, e.g. NY -> 0, WA -> 1?
- Answer: There is no inherent ordering to states, e.g. WA is not inherently "more" of anything than NY.
- We've already seen the correct strategy: one hot encoding.
Example: Horsepower 🚗¶
The following dataset, built into the seaborn plotting library, contains various information about (older) cars.
mpg = sns.load_dataset('mpg').dropna()
mpg.head()
| mpg | cylinders | displacement | horsepower | ... | acceleration | model_year | origin | name | |
|---|---|---|---|---|---|---|---|---|---|
| 0 | 18.0 | 8 | 307.0 | 130.0 | ... | 12.0 | 70 | usa | chevrolet chevelle malibu |
| 1 | 15.0 | 8 | 350.0 | 165.0 | ... | 11.5 | 70 | usa | buick skylark 320 |
| 2 | 18.0 | 8 | 318.0 | 150.0 | ... | 11.0 | 70 | usa | plymouth satellite |
| 3 | 16.0 | 8 | 304.0 | 150.0 | ... | 12.0 | 70 | usa | amc rebel sst |
| 4 | 17.0 | 8 | 302.0 | 140.0 | ... | 10.5 | 70 | usa | ford torino |
5 rows × 9 columns
We really do mean old:
mpg['model_year'].value_counts()
73 40
78 36
76 34
..
71 27
80 27
74 26
Name: model_year, Length: 13, dtype: int64
Let's investigate the relationship between 'horsepower' and 'mpg'.
The relationship between 'horsepower' and 'mpg'¶
px.scatter(mpg, x='horsepower', y='mpg')
- It appears that there is a negative association between
'horsepower'and'mpg', though it's not quite linear.
- Let's try and fit a simple linear model that uses
'horsepower'to predict'mpg'and see what happens.
Predicting 'mpg' using 'horsepower'¶
car_model = LinearRegression()
car_model.fit(mpg[['horsepower']], mpg['mpg'])
LinearRegression()
What do our predictions look like?
hp_points = pd.DataFrame({'horsepower': [25, 225]})
fig = px.scatter(mpg, x='horsepower', y='mpg')
fig.add_trace(go.Scatter(
x=hp_points['horsepower'],
y=car_model.predict(hp_points),
mode='lines',
name='Predicted MPG using Horsepower'
))
Our regression line doesn't capture the curvature in the relationship between 'horsepower' and 'mpg'.
Let's look at the residuals:
res = mpg.assign(
Predictions=car_model.predict(mpg[['horsepower']]),
Residuals=mpg['mpg'] - car_model.predict(mpg[['horsepower']]),
)
fig = px.scatter(res, x='Predictions', y='Residuals')
fig.add_hline(0, line_width=3, opacity=1)
car_model.score(mpg[['horsepower']], mpg['mpg'])
0.6059482578894348
Linearization¶
The Tukey Mosteller Bulge Diagram helps us pick which transformations to apply to data in order to linearize it.
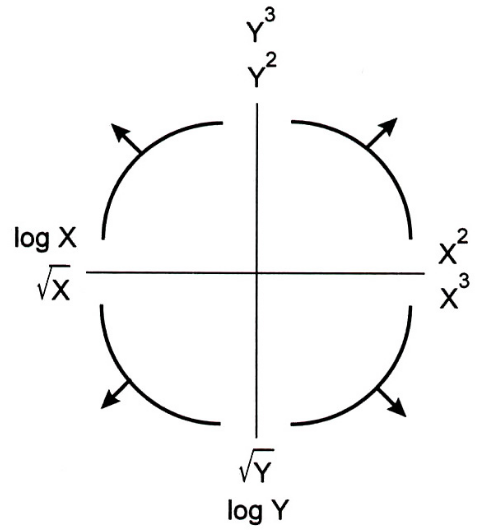
The bottom-left quadrant appears to match the shape of the scatter plot between 'horsepower' and 'mpg' the best – let's try taking the log of 'horsepower' ($X$).
mpg['log hp'] = np.log(mpg['horsepower'])
What does our data look like now?
px.scatter(mpg, x='log hp', y='mpg')
Predicting 'mpg' using log('horsepower')¶
Let's fit another linear model.
car_model_log = LinearRegression()
car_model_log.fit(mpg[['log hp']], mpg['mpg'])
LinearRegression()
What do our predictions look like now?
fig = px.scatter(mpg, x='log hp', y='mpg')
log_hp_points = pd.DataFrame({'log hp': [3.7, 5.5]})
fig = px.scatter(mpg, x='log hp', y='mpg')
fig.add_trace(go.Scatter(
x=log_hp_points['log hp'],
y=car_model_log.predict(log_hp_points),
mode='lines',
name='Predicted MPG using log(Horsepower)'
))
The fit looks a bit better! How about the $R^2$?
car_model_log.score(mpg[['log hp']], mpg['mpg'])
0.6683347641192137
Also a bit better!
What do our predictions look like on the original, non-transformed scatter plot? Let's see:
fig = px.scatter(mpg, x='horsepower', y='mpg')
fig.add_trace(
go.Scatter(
x=mpg['horsepower'],
y=car_model_log.intercept_ + car_model_log.coef_[0] * np.log(mpg['horsepower']),
mode='markers', name='Predicted MPG using log(Horsepower)'
)
)
fig
Our predictions that used $\log(\text{Horsepower})$ as an input don't fall on a straight line. We shouldn't expect them to; the red dots come from:
$$\text{Predicted MPG} = 108.698 - 18.582 \cdot \log(\text{Horsepower})$$car_model_log.intercept_, car_model_log.coef_
(108.69970699574486, array([-18.58]))
Quantitative scaling¶
Until now, feature transformations we've discussed so far have involved converting categorical variables into quantitative variables. However, our log transformation was an example of transforming a quantitative variable into a new quantitative variable; this practice is called quantitative scaling.
- Standardization: $x_i \rightarrow \frac{x_i - \bar{x}}{\sigma_x}$.
- Linearization via a non-linear transformation: e.g. $\text{log}$ and $\text{sqrt}$. See Lab 8 for more.
- Discretization: Convert data into percentiles (or more generally, quantiles).
Question 🤔 (Answer at q.dsc80.com)
What questions do you have?
The modeling process¶
The modeling process¶
- Create, or engineer, features to best reflect the "meaning" behind data.
- Choose a model that is appropriate to capture the relationships between features ($X$) and the target/response ($y$).
- Choose a loss function, e.g. squared loss.
- Fit the model: that is, minimize empirical risk to find optimal model parameters $w^*$.
- Evaluate the model, e.g. using RMSE or $R^2$.
We can perform all of the above directly in sklearn!

preprocessing and linear_models¶
For the feature engineering step of the modeling pipeline, we will use sklearn's preprocessing module.
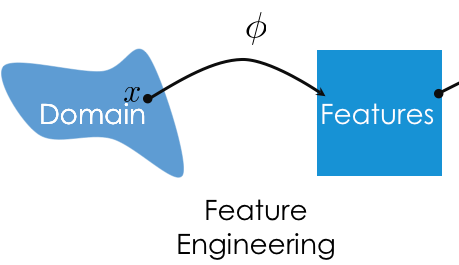
For the model creation step of the modeling pipeline, we will use sklearn's linear_model module, as we've already seen. linear_model.LinearRegression is an example of an estimator class.
Transformers in sklearn¶
Transformer classes¶
- Transformers take in "raw" data and output "processed" data. They are used for creating features.
- The input to a transformer should be a multi-dimensional
numpyarray.- Inputs can be DataFrames, but
sklearnonly looks at the values (i.e. it callsto_numpy()on input DataFrames).
- Inputs can be DataFrames, but
- The output of a transformer is a
numpyarray (never a DataFrame or Series).
- Transformers, like most relevant features of
sklearn, are classes, not functions, meaning you need to instantiate them and call their methods.
Example: Predicting tips 🧑🍳¶
We'll continue working with our trusty tips dataset.
tips.head()
| total_bill | tip | sex | smoker | ... | time | size | smoker == Yes | smoker == No | |
|---|---|---|---|---|---|---|---|---|---|
| 0 | 3.07 | 1.00 | Female | Yes | ... | Dinner | 1 | 1 | 0 |
| 1 | 18.78 | 3.00 | Female | No | ... | Dinner | 2 | 0 | 1 |
| 2 | 26.59 | 3.41 | Male | Yes | ... | Dinner | 3 | 1 | 0 |
| 3 | 14.26 | 2.50 | Male | No | ... | Lunch | 2 | 0 | 1 |
| 4 | 21.16 | 3.00 | Male | No | ... | Lunch | 2 | 0 | 1 |
5 rows × 9 columns
Example transformer: Binarizer¶
The Binarizer transformer allows us to map a quantitative sequence to a sequence of 1s and 0s, depending on whether values are above or below a threshold.
| Property | Example | Description |
|---|---|---|
| Initialize with parameters | binar = Binarizer(thresh) |
set x=1 if x > thresh, else 0 |
| Transform data in a dataset | feat = binar.transform(data) |
Binarize all columns in data |
First, we need to import the relevant class from sklearn.preprocessing. (Tip: import just the relevant classes you need from sklearn.)
from sklearn.preprocessing import Binarizer
Let's try binarizing 'total_bill'. We'll say a "large" bill is one that is strictly greater than $20.
tips['total_bill'].head()
0 3.07 1 18.78 2 26.59 3 14.26 4 21.16 Name: total_bill, dtype: float64
First, we initialize a Binarizer object with the threshold we want.
bi = Binarizer(threshold=20)
Then, we call bi's transform method and pass it the data we'd like to transform. Note that its input and output are both 2D.
transformed_bills = bi.transform(tips[['total_bill']]) # Must give transform a 2D array/DataFrame.
transformed_bills[:5]
/Users/sam/mambaforge/envs/dsc80/lib/python3.8/site-packages/sklearn/base.py:443: UserWarning: X has feature names, but Binarizer was fitted without feature names
array([[0.],
[0.],
[1.],
[0.],
[1.]])
Example transformer: StandardScaler¶
StandardScalerstandardizes data using the mean and standard deviation of the data.
- Unlike
Binarizer,StandardScalerrequires some knowledge (mean and SD) of the dataset before transforming.
- As such, we need to
fitanStandardScalertransformer before we can use thetransformmethod.
- Typical usage: fit transformer on a sample, use that fit transformer to transform future data.
Example transformer: StandardScaler¶
It only makes sense to standardize the already-quantitative features of tips, so let's select just those.
tips_quant = tips[['total_bill', 'size']]
tips_quant.head()
| total_bill | size | |
|---|---|---|
| 0 | 3.07 | 1 |
| 1 | 18.78 | 2 |
| 2 | 26.59 | 3 |
| 3 | 14.26 | 2 |
| 4 | 21.16 | 2 |
Let's initialize a StandardScaler object.
from sklearn.preprocessing import StandardScaler
stdscaler = StandardScaler()
Note that the following does not work! The error message is very helpful.
stdscaler.transform(tips_quant)
--------------------------------------------------------------------------- NotFittedError Traceback (most recent call last) Cell In[50], line 1 ----> 1 stdscaler.transform(tips_quant) File ~/mambaforge/envs/dsc80/lib/python3.8/site-packages/sklearn/preprocessing/_data.py:970, in StandardScaler.transform(self, X, copy) 955 def transform(self, X, copy=None): 956 """Perform standardization by centering and scaling. 957 958 Parameters (...) 968 Transformed array. 969 """ --> 970 check_is_fitted(self) 972 copy = copy if copy is not None else self.copy 973 X = self._validate_data( 974 X, 975 reset=False, (...) 980 force_all_finite="allow-nan", 981 ) File ~/mambaforge/envs/dsc80/lib/python3.8/site-packages/sklearn/utils/validation.py:1222, in check_is_fitted(estimator, attributes, msg, all_or_any) 1217 fitted = [ 1218 v for v in vars(estimator) if v.endswith("_") and not v.startswith("__") 1219 ] 1221 if not fitted: -> 1222 raise NotFittedError(msg % {"name": type(estimator).__name__}) NotFittedError: This StandardScaler instance is not fitted yet. Call 'fit' with appropriate arguments before using this estimator.
Instead, we need to first call the fit method on stdscaler.
# This is like saying "determine the mean and SD of each column in tips_quant".
stdscaler.fit(tips_quant)
StandardScaler()
Now, transform will work.
# First column is 'total_bill', second column is 'size'.
tips_quant_z = stdscaler.transform(tips_quant)
tips_quant_z[:5]
array([[-1.88, -1.65],
[-0.11, -0.6 ],
[ 0.77, 0.45],
[-0.62, -0.6 ],
[ 0.15, -0.6 ]])
We can also access the mean and variance stdscaler computed for each column:
stdscaler.mean_
array([19.79, 2.57])
stdscaler.var_
array([78.93, 0.9 ])
Note that we can call transform on DataFrames other than tips_quant. We will do this often – fit a transformer on one dataset (training data) and use it to transform other datasets (test data).
stdscaler.transform(tips_quant.sample(5))
array([[ 0.97, -0.6 ],
[-0.28, -0.6 ],
[ 1.19, 1.51],
[-0.49, -0.6 ],
[ 0.65, 1.51]])
💡 Pro-Tip: Using .fit_transform¶
The .fit_transform method will fit the transformer and then transform the data in one go.
stdscaler.fit_transform(tips_quant)
array([[-1.88, -1.65],
[-0.11, -0.6 ],
[ 0.77, 0.45],
...,
[-0.26, -0.6 ],
[-1.09, -0.6 ],
[-0.32, 0.45]])
StandardScaler summary¶
| Property | Example | Description |
|---|---|---|
| Initialize with parameters | stdscaler = StandardScaler() |
z-score the data (no parameters) |
| Fit the transformer | stdscaler.fit(X) |
Compute the mean and SD of X |
| Transform data in a dataset | feat = stdscaler.transform(X_new) |
z-score X_new with mean and SD of X |
| Fit and transform | stdscaler.fit_transform(X) |
Compute the mean and SD of X, then z-score X |
Example transformer: OneHotEncoder¶
Let's keep just the categorical columns in tips.
tips_cat = tips[['sex', 'smoker', 'day', 'time']]
tips_cat.head()
| sex | smoker | day | time | |
|---|---|---|---|---|
| 0 | Female | Yes | Sat | Dinner |
| 1 | Female | No | Thur | Dinner |
| 2 | Male | Yes | Sat | Dinner |
| 3 | Male | No | Thur | Lunch |
| 4 | Male | No | Thur | Lunch |
Like StdScaler, we will need to fit our OneHotEncoder transformer before it can transform anything.
from sklearn.preprocessing import OneHotEncoder
ohe = OneHotEncoder()
ohe.fit(tips_cat)
OneHotEncoder()
When we try and transform, we get a result we might not expect.
ohe.transform(tips_cat)
<244x10 sparse matrix of type '<class 'numpy.float64'>' with 976 stored elements in Compressed Sparse Row format>
Since the resulting matrix is sparse – most of its elements are 0 – sklearn uses a more efficient representation than a regular numpy array. We can convert to a regular (dense) array:
ohe.transform(tips_cat).toarray()
array([[1., 0., 0., ..., 0., 1., 0.],
[1., 0., 1., ..., 1., 1., 0.],
[0., 1., 0., ..., 0., 1., 0.],
...,
[1., 0., 1., ..., 1., 0., 1.],
[0., 1., 1., ..., 0., 1., 0.],
[1., 0., 1., ..., 0., 1., 0.]])
Notice that the column names from tips_cat are no longer stored anywhere (remember, fit converts the input to a numpy array before proceeding).
We can use the get_feature_names_out method on ohe to access the names of the one-hot-encoded columns, though:
ohe.get_feature_names_out() # x0, x1, x2, and x3 correspond to column names in tips_cat.
array(['sex_Female', 'sex_Male', 'smoker_No', 'smoker_Yes', 'day_Fri',
'day_Sat', 'day_Sun', 'day_Thur', 'time_Dinner', 'time_Lunch'],
dtype=object)
pd.DataFrame(ohe.transform(tips_cat).toarray(),
columns=ohe.get_feature_names_out()) # If we need a DataFrame back, for some reason.
| sex_Female | sex_Male | smoker_No | smoker_Yes | ... | day_Sun | day_Thur | time_Dinner | time_Lunch | |
|---|---|---|---|---|---|---|---|---|---|
| 0 | 1.0 | 0.0 | 0.0 | 1.0 | ... | 0.0 | 0.0 | 1.0 | 0.0 |
| 1 | 1.0 | 0.0 | 1.0 | 0.0 | ... | 0.0 | 1.0 | 1.0 | 0.0 |
| 2 | 0.0 | 1.0 | 0.0 | 1.0 | ... | 0.0 | 0.0 | 1.0 | 0.0 |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 241 | 1.0 | 0.0 | 1.0 | 0.0 | ... | 0.0 | 1.0 | 0.0 | 1.0 |
| 242 | 0.0 | 1.0 | 1.0 | 0.0 | ... | 0.0 | 0.0 | 1.0 | 0.0 |
| 243 | 1.0 | 0.0 | 1.0 | 0.0 | ... | 0.0 | 0.0 | 1.0 | 0.0 |
244 rows × 10 columns
Question 🤔 (Answer at q.dsc80.com)
Suppose we have a training set X_train with labels y_train, and we have a test set X_test with labels y_test. If we want to use OHE on the day column, we will use ohe.fit_transform(X_train), but we will NOT do ohe.fit_transform(X_test). Instead, we will ONLY do ohe.transform(X_test). Why?
We ended lecture here.
Pipelines¶

So far, we've used transformers for feature engineering and models for prediction. We can combine these steps into a single Pipeline.
Pipelines in sklearn¶
From sklearn's documentation:
Pipeline allows you to sequentially apply a list of transformers to preprocess the data and, if desired, conclude the sequence with a final predictor for predictive modeling.
Intermediate steps of the pipeline must be "transforms", that is, they must implementfitandtransformmethods. The final estimator only needs to implementfit.
- General template:
pl = Pipeline([trans_1, trans_2, ..., model])- Note that the
modelis optional.
- Note that the
- Once a
Pipelineis instantiated, you can fit all steps (transformers and model) usingpl.fit(X, y).
- To make predictions using raw, untransformed data, use
pl.predict(X).
- The actual list we provide
Pipelinewith must be a list of tuples, where- The first element is a "name" (that we choose) for the step.
- The second element is a transformer or estimator instance.
Our first Pipeline¶
Let's build a Pipeline that:
- One hot encodes the categorical features in
tips. - Fits a regression model on the one hot encoded data.
tips_cat = tips[['sex', 'smoker', 'day', 'time']]
tips_cat.head()
| sex | smoker | day | time | |
|---|---|---|---|---|
| 0 | Female | Yes | Sat | Dinner |
| 1 | Female | No | Thur | Dinner |
| 2 | Male | Yes | Sat | Dinner |
| 3 | Male | No | Thur | Lunch |
| 4 | Male | No | Thur | Lunch |
from sklearn.pipeline import Pipeline
pl = Pipeline([
('one-hot', OneHotEncoder()),
('lin-reg', LinearRegression())
])
Now that pl is instantiated, we fit it the same way we would fit the individual steps.
pl.fit(tips_cat, tips['tip'])
Pipeline(steps=[('one-hot', OneHotEncoder()), ('lin-reg', LinearRegression())])
Now, to make predictions using raw data, all we need to do is use pl.predict:
pl.predict(tips_cat.iloc[:5])
array([2.92, 3.16, 3.09, 2.83, 2.83])
pl performs both feature transformation and prediction with just a single call to predict!
We can access individual "steps" of a Pipeline through the named_steps attribute:
pl.named_steps
{'one-hot': OneHotEncoder(), 'lin-reg': LinearRegression()}
pl.named_steps['one-hot'].transform(tips_cat).toarray()
array([[1., 0., 0., ..., 0., 1., 0.],
[1., 0., 1., ..., 1., 1., 0.],
[0., 1., 0., ..., 0., 1., 0.],
...,
[1., 0., 1., ..., 1., 0., 1.],
[0., 1., 1., ..., 0., 1., 0.],
[1., 0., 1., ..., 0., 1., 0.]])
pl.named_steps['one-hot'].get_feature_names_out()
array(['sex_Female', 'sex_Male', 'smoker_No', 'smoker_Yes', 'day_Fri',
'day_Sat', 'day_Sun', 'day_Thur', 'time_Dinner', 'time_Lunch'],
dtype=object)
pl.named_steps['lin-reg'].coef_
array([-0.09, 0.09, -0.04, 0.04, -0.2 , -0.13, 0.14, 0.19, 0.25,
-0.25])
pl also has a score method, the same way a fit LinearRegression instance does:
# Why is this so low?
pl.score(tips_cat, tips['tip'])
0.02749679020147533
More sophisticated Pipelines¶
- In the previous example, we one hot encoded every input column. What if we want to perform different transformations on different columns?
- Solution: Use a
ColumnTransformer.- Instantiate a
ColumnTransformerusing a list of tuples, where:- The first element is a "name" we choose for the transformer.
- The second element is a transformer instance (e.g.
OneHotEncoder()). - The third element is a list of relevant column names.
- Instantiate a
Planning our first ColumnTransformer¶
from sklearn.compose import ColumnTransformer
Let's perform different transformations on the quantitative and categorical features of tips (note that we are not transforming 'tip').
tips_features = tips.drop('tip', axis=1)
tips_features.head()
| total_bill | sex | smoker | day | time | size | smoker == Yes | smoker == No | |
|---|---|---|---|---|---|---|---|---|
| 0 | 3.07 | Female | Yes | Sat | Dinner | 1 | 1 | 0 |
| 1 | 18.78 | Female | No | Thur | Dinner | 2 | 0 | 1 |
| 2 | 26.59 | Male | Yes | Sat | Dinner | 3 | 1 | 0 |
| 3 | 14.26 | Male | No | Thur | Lunch | 2 | 0 | 1 |
| 4 | 21.16 | Male | No | Thur | Lunch | 2 | 0 | 1 |
- We will leave the
'total_bill'column untouched.
- To the
'size'column, we will apply theBinarizertransformer with a threshold of 2 (big tables vs. small tables).
- To the categorical columns, we will apply the
OneHotEncodertransformer.
- In essence, we will create a transformer that reproduces the following DataFrame:
| size | x0_Female | x0_Male | x1_No | x1_Yes | x2_Fri | x2_Sat | x2_Sun | x2_Thur | x3_Dinner | x3_Lunch | total_bill | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 0 | 1.0 | 0.0 | 1.0 | 0.0 | 0.0 | 0.0 | 1.0 | 0.0 | 1.0 | 0.0 | 16.99 |
| 1 | 1 | 0.0 | 1.0 | 1.0 | 0.0 | 0.0 | 0.0 | 1.0 | 0.0 | 1.0 | 0.0 | 10.34 |
| 2 | 1 | 0.0 | 1.0 | 1.0 | 0.0 | 0.0 | 0.0 | 1.0 | 0.0 | 1.0 | 0.0 | 21.01 |
| 3 | 0 | 0.0 | 1.0 | 1.0 | 0.0 | 0.0 | 0.0 | 1.0 | 0.0 | 1.0 | 0.0 | 23.68 |
| 4 | 1 | 1.0 | 0.0 | 1.0 | 0.0 | 0.0 | 0.0 | 1.0 | 0.0 | 1.0 | 0.0 | 24.59 |
Building a Pipeline using a ColumnTransformer¶
Let's start by creating our ColumnTransformer.
preproc = ColumnTransformer(
transformers=[
('size', Binarizer(threshold=2), ['size']),
('categorical_cols', OneHotEncoder(), ['sex', 'smoker', 'day', 'time'])
],
remainder='passthrough' # Specify what to do with all other columns ('total_bill' here) – drop or passthrough.
)
Now, let's create a Pipeline using preproc as a transformer, and fit it:
pl = Pipeline([
('preprocessor', preproc),
('lin-reg', LinearRegression())
])
pl.fit(tips_features, tips['tip'])
Pipeline(steps=[('preprocessor',
ColumnTransformer(remainder='passthrough',
transformers=[('size',
Binarizer(threshold=2),
['size']),
('categorical_cols',
OneHotEncoder(),
['sex', 'smoker', 'day',
'time'])])),
('lin-reg', LinearRegression())])
Prediction is as easy as calling predict:
tips_features.head()
| total_bill | sex | smoker | day | time | size | smoker == Yes | smoker == No | |
|---|---|---|---|---|---|---|---|---|
| 0 | 3.07 | Female | Yes | Sat | Dinner | 1 | 1 | 0 |
| 1 | 18.78 | Female | No | Thur | Dinner | 2 | 0 | 1 |
| 2 | 26.59 | Male | Yes | Sat | Dinner | 3 | 1 | 0 |
| 3 | 14.26 | Male | No | Thur | Lunch | 2 | 0 | 1 |
| 4 | 21.16 | Male | No | Thur | Lunch | 2 | 0 | 1 |
# Note that we fit the Pipeline using tips_features, not tips_features.head()!
pl.predict(tips_features.head())
array([1.16, 2.81, 3.7 , 2.4 , 3.07])
Aside: FunctionTransformer¶
A transformer you'll often use as part of a ColumnTransformer is the FunctionTransformer, which enables you to use your own functions on entire columns. Think of it as the sklearn equivalent of apply.
from sklearn.preprocessing import FunctionTransformer
f = FunctionTransformer(np.sqrt)
f.transform([1, 2, 3])
array([1. , 1.41, 1.73])
Summary, next time¶
Summary¶
- To transform a categorical nominal variable into a quantitative variable, use one hot encoding.
- To transform a categorical ordinal variable into a quantitative variable, use an ordinal encoding.
- Quantitative feature transformations allow us to use linear models to model non-linear data.
Pipelines are powerful because they allow you to perform feature engineering and training/prediction all through a single object.- As we'll see next time, they also allow us to easily compare models against each other.
Next time¶
- More examples of, and ways to create,
Pipelines. - Multicollinearity.
- Generalization.