# You'll start seeing this cell in most lectures.
# It exists to hide all of the import statements and other setup
# code we need in lecture notebooks.
from dsc80_utils import *
In this reading, we'll review some of the basics of numpy and babypandas that you're familiar with from DSC 10. In lecture, we'll build off of this foundation.
numpy arrays¶
numpy overview¶
numpystands for "numerical Python". It is a commonly-used Python module that enables fast computation involving arrays and matrices.numpy's main object is the array. Innumpy, arrays are:- Homogenous – all values are of the same type.
- (Potentially) multi-dimensional.
- Computation in
numpyis fast because:- Much of it is implemented in C.
numpyarrays are stored more efficiently in memory than, say, Python lists.
- This site provides a good overview of
numpyarrays.
We used numpy in DSC 10 to work with sequences of data:
arr = np.arange(10)
arr
array([0, 1, 2, 3, 4, 5, 6, 7, 8, 9])
# The shape (10,) means that the array only has a single dimension,
# of size 10.
arr.shape
(10,)
2 ** arr
array([ 1, 2, 4, 8, 16, 32, 64, 128, 256, 512])
Arrays come equipped with several handy methods; some examples are below, but you can read about them all here.
(2 ** arr).sum()
1023
(2 ** arr).mean()
102.3
(2 ** arr).max()
512
(2 ** arr).argmax()
9
⚠️ The dangers of for-loops¶
for-loops are slow when processing large datasets. You will rarely writefor-loops in DSC 80 (except for Lab 1 and Project 1), and may be penalized on assignments for using them when unnecessary!- One of the biggest benefits of
numpyis that it supports vectorized operations.- If
aandbare two arrays of the same length, thena + bis a new array of the same length containing the element-wise sum ofaandb.
- If
- To illustrate how much faster
numpyarithmetic is than using afor-loop, let's compute the squares of the numbers between 0 and 1,000,000:- Using a
for-loop. - Using vectorized arithmetic, through
numpy.
- Using a
%%timeit
squares = []
for i in range(1_000_000):
squares.append(i * i)
47.6 ms ± 526 µs per loop (mean ± std. dev. of 7 runs, 10 loops each)
In vanilla Python, this takes about 0.04 seconds per loop.
%%timeit
squares = np.arange(1_000_000) ** 2
1.46 ms ± 77.1 µs per loop (mean ± std. dev. of 7 runs, 1,000 loops each)
In numpy, this only takes about 0.001 seconds per loop, more than 40x faster! Note that under the hood, numpy is also using a for-loop, but it's a for-loop implemented in C, which is much faster than Python.
Multi-dimensional arrays¶
While we didn't see these very often in DSC 10, multi-dimensional lists/arrays may have since come up in DSC 20, 30, or 40A (especially in the context of linear algebra).
We'll spend a bit of time talking about 2D (and 3D) arrays here, since in some ways, they behave similarly to DataFrames.
Below, we create a 2D array from scratch.
nums = np.array([
[5, 1, 9, 7],
[9, 8, 2, 3],
[2, 5, 0, 4]
])
nums
array([[5, 1, 9, 7],
[9, 8, 2, 3],
[2, 5, 0, 4]])
# nums has 3 rows and 4 columns.
nums.shape
(3, 4)
We can also create 2D arrays by reshaping other arrays.
# Here, we're asking to reshape np.arange(1, 7)
# so that it has 2 rows and 3 columns.
a = np.arange(1, 7).reshape((2, 3))
a
array([[1, 2, 3],
[4, 5, 6]])
Operations along axes¶
In 2D arrays (and DataFrames), axis 0 refers to the rows (up and down) and axis 1 refers to the columns (left and right).
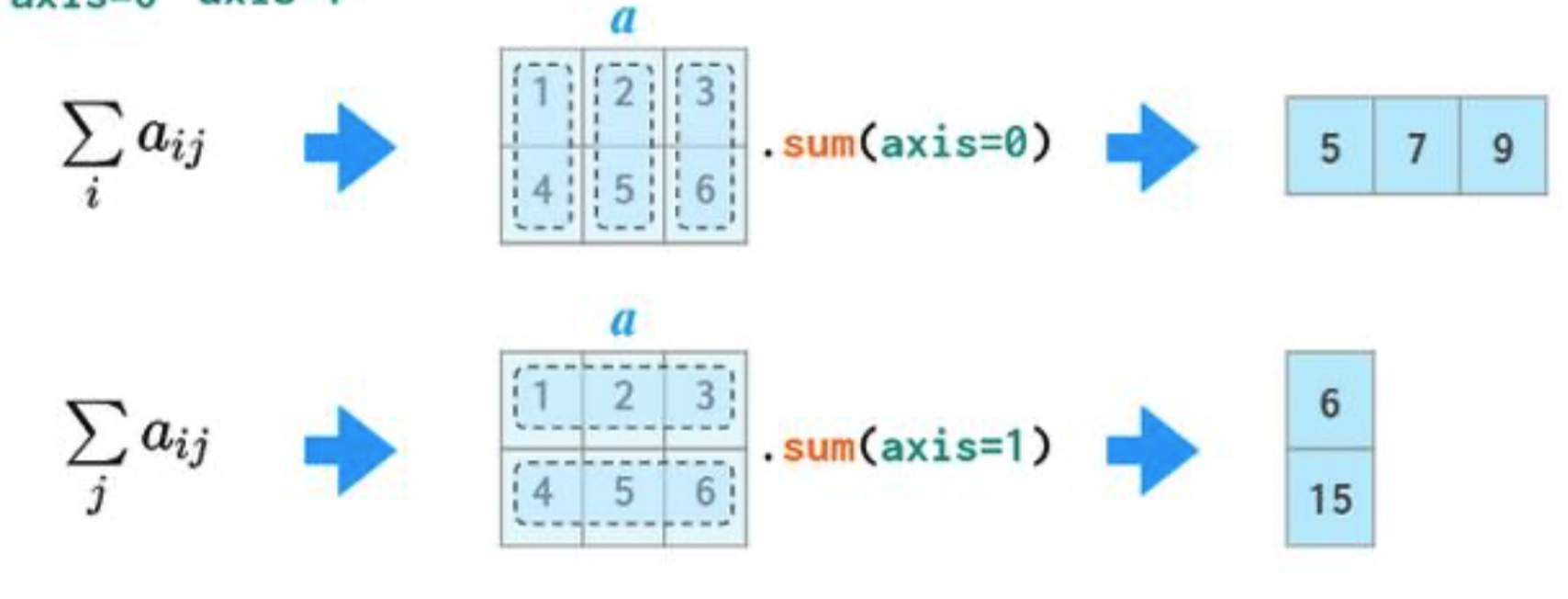
a
array([[1, 2, 3],
[4, 5, 6]])
If we specify axis=0, a.sum will "compress" along axis 0.
a.sum(axis=0)
array([5, 7, 9])
If we specify axis=1, a.sum will "compress" along axis 1.
a.sum(axis=1)
array([ 6, 15])
Selecting rows and columns from 2D arrays¶
You can use [square brackets] to slice rows and columns out of an array, using the same slicing conventions you saw in DSC 20.
a
array([[1, 2, 3],
[4, 5, 6]])
# Accesses row 0 and all columns.
a[0, :]
array([1, 2, 3])
# Same as the above.
a[0]
array([1, 2, 3])
# Accesses all rows and column 1.
a[:, 1]
array([2, 5])
# Accesses row 0 and columns 1 and onwards.
a[0, 1:]
array([2, 3])
Exercise
Try and predict the value ofgrid[-1, 1:].sum() without running the code below.
s = (5, 3)
grid = np.ones(s) * 2 * np.arange(1, 16).reshape(s)
# grid[-1, 1:].sum()
From babypandas to pandas 🐼¶
babypandas¶
In DSC 10, you used babypandas, which was a subset of pandas designed to be friendly for beginners.

pandas¶
You're not a beginner anymore – you've taken DSC 20, 30, and 40A. You're ready for the real deal.

Fortunately, everything you learned in babypandas will carry over!
pandas¶

pandasis the Python library for tabular data manipulation.- Before
pandaswas developed, the standard data science workflow involved using multiple languages (Python, R, Java) in a single project. - Wes McKinney, the original developer of
pandas, wanted a library which would allow everything to be done in Python.- Python is faster to develop in than Java, and is more general-purpose than R.
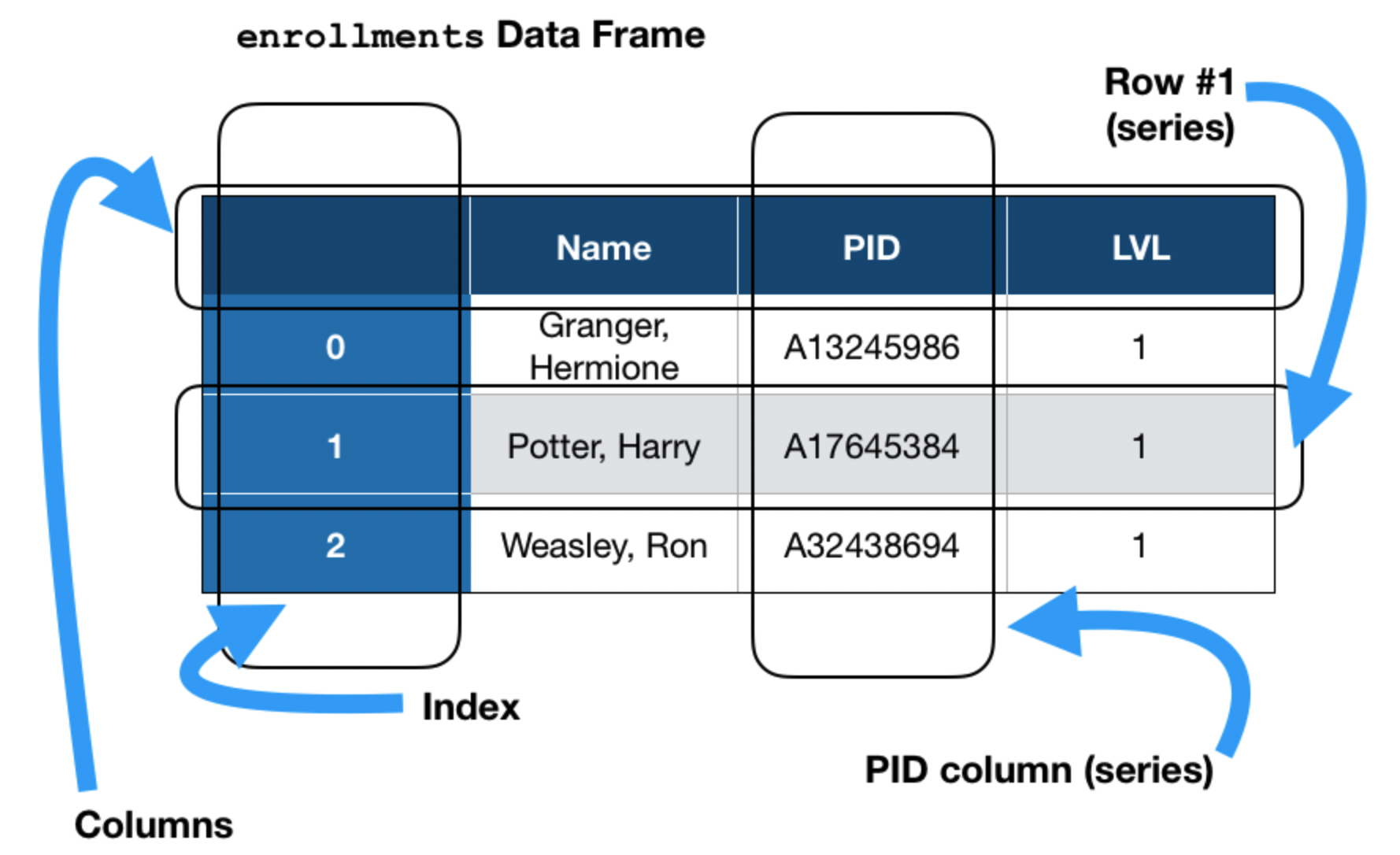
pandas data structures¶
There are three key data structures at the core of pandas:
- DataFrame: 2 dimensional tables.
- Series: 1 dimensional array-like object, typically representing a column or row.
- Index: sequence of column or row labels.
Importing pandas and related libraries¶
pandas is almost always imported in conjunction with numpy.
import pandas as pd
import numpy as np
Example: Dog Breeds (woof!) 🐶¶
We'll provide more context for the dataset we're working with in lecture. For now, all you need to know is that each row corresponds to a different dog breed.
# You'll see the Path(...) / syntax a lot.
# It creates the correct path to your file,
# whether you're using Windows, macOS, or Linux.
# (Note that macOS and Linux use / to denote separate folders in paths,
# while Windows uses \.)
dog_path = Path('data') / 'dogs43.csv'
dogs = pd.read_csv(dog_path)
dogs
| breed | kind | lifetime_cost | longevity | size | weight | height | |
|---|---|---|---|---|---|---|---|
| 0 | Brittany | sporting | 22589.0 | 12.92 | medium | 35.0 | 19.0 |
| 1 | Cairn Terrier | terrier | 21992.0 | 13.84 | small | 14.0 | 10.0 |
| 2 | English Cocker Spaniel | sporting | 18993.0 | 11.66 | medium | 30.0 | 16.0 |
| ... | ... | ... | ... | ... | ... | ... | ... |
| 40 | Bullmastiff | working | 13936.0 | 7.57 | large | 115.0 | 25.5 |
| 41 | Mastiff | working | 13581.0 | 6.50 | large | 175.0 | 30.0 |
| 42 | Saint Bernard | working | 20022.0 | 7.78 | large | 155.0 | 26.5 |
43 rows × 7 columns
Review: head, tail, shape, index, get, and sort_values¶
To extract the first or last few rows of a DataFrame, use the head or tail methods.
dogs.head(3)
| breed | kind | lifetime_cost | longevity | size | weight | height | |
|---|---|---|---|---|---|---|---|
| 0 | Brittany | sporting | 22589.0 | 12.92 | medium | 35.0 | 19.0 |
| 1 | Cairn Terrier | terrier | 21992.0 | 13.84 | small | 14.0 | 10.0 |
| 2 | English Cocker Spaniel | sporting | 18993.0 | 11.66 | medium | 30.0 | 16.0 |
dogs.tail(2)
| breed | kind | lifetime_cost | longevity | size | weight | height | |
|---|---|---|---|---|---|---|---|
| 41 | Mastiff | working | 13581.0 | 6.50 | large | 175.0 | 30.0 |
| 42 | Saint Bernard | working | 20022.0 | 7.78 | large | 155.0 | 26.5 |
The shape attribute returns the DataFrame's number of rows and columns.
dogs.shape
(43, 7)
# The default index of a DataFrame is 0, 1, 2, 3, ...
dogs.index
RangeIndex(start=0, stop=43, step=1)
We know that we can use .get() to select out a column or multiple columns...
dogs.get('breed')
0 Brittany
1 Cairn Terrier
2 English Cocker Spaniel
...
40 Bullmastiff
41 Mastiff
42 Saint Bernard
Name: breed, Length: 43, dtype: object
dogs.get(['breed', 'kind', 'longevity'])
| breed | kind | longevity | |
|---|---|---|---|
| 0 | Brittany | sporting | 12.92 |
| 1 | Cairn Terrier | terrier | 13.84 |
| 2 | English Cocker Spaniel | sporting | 11.66 |
| ... | ... | ... | ... |
| 40 | Bullmastiff | working | 7.57 |
| 41 | Mastiff | working | 6.50 |
| 42 | Saint Bernard | working | 7.78 |
43 rows × 3 columns
Most people don't use .get in practice; we'll see the more common technique in lecture.
And lastly, remember that to sort by a column, use the sort_values method. Like most DataFrame and Series methods, sort_values returns a new DataFrame, and doesn't modify the original.
# Note that the index is no longer 0, 1, 2, ...!
dogs.sort_values('height', ascending=False)
| breed | kind | lifetime_cost | longevity | size | weight | height | |
|---|---|---|---|---|---|---|---|
| 41 | Mastiff | working | 13581.0 | 6.50 | large | 175.0 | 30.0 |
| 36 | Borzoi | hound | 16176.0 | 9.08 | large | 82.5 | 28.0 |
| 34 | Newfoundland | working | 19351.0 | 9.32 | large | 125.0 | 27.0 |
| ... | ... | ... | ... | ... | ... | ... | ... |
| 29 | Dandie Dinmont Terrier | terrier | 21633.0 | 12.17 | small | 21.0 | 9.0 |
| 14 | Maltese | toy | 19084.0 | 12.25 | small | 5.0 | 9.0 |
| 8 | Chihuahua | toy | 26250.0 | 16.50 | small | 5.5 | 5.0 |
43 rows × 7 columns
# This sorts by 'height',
# then breaks ties by 'longevity'.
# Note the difference in the last three rows between
# this DataFrame and the one above.
dogs.sort_values(['height', 'longevity'],
ascending=False)
| breed | kind | lifetime_cost | longevity | size | weight | height | |
|---|---|---|---|---|---|---|---|
| 41 | Mastiff | working | 13581.0 | 6.50 | large | 175.0 | 30.0 |
| 36 | Borzoi | hound | 16176.0 | 9.08 | large | 82.5 | 28.0 |
| 34 | Newfoundland | working | 19351.0 | 9.32 | large | 125.0 | 27.0 |
| ... | ... | ... | ... | ... | ... | ... | ... |
| 14 | Maltese | toy | 19084.0 | 12.25 | small | 5.0 | 9.0 |
| 29 | Dandie Dinmont Terrier | terrier | 21633.0 | 12.17 | small | 21.0 | 9.0 |
| 8 | Chihuahua | toy | 26250.0 | 16.50 | small | 5.5 | 5.0 |
43 rows × 7 columns
Note that dogs is not the DataFrame above. To save our changes, we'd need to say something like dogs = dogs.sort_values....
dogs
| breed | kind | lifetime_cost | longevity | size | weight | height | |
|---|---|---|---|---|---|---|---|
| 0 | Brittany | sporting | 22589.0 | 12.92 | medium | 35.0 | 19.0 |
| 1 | Cairn Terrier | terrier | 21992.0 | 13.84 | small | 14.0 | 10.0 |
| 2 | English Cocker Spaniel | sporting | 18993.0 | 11.66 | medium | 30.0 | 16.0 |
| ... | ... | ... | ... | ... | ... | ... | ... |
| 40 | Bullmastiff | working | 13936.0 | 7.57 | large | 115.0 | 25.5 |
| 41 | Mastiff | working | 13581.0 | 6.50 | large | 175.0 | 30.0 |
| 42 | Saint Bernard | working | 20022.0 | 7.78 | large | 155.0 | 26.5 |
43 rows × 7 columns
That's all we need to review... we'll pick back up in lecture!